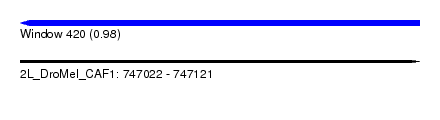
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 747,022 – 747,121 |
| Length | 99 |
| Max. P | 0.977181 |

| Location | 747,022 – 747,121 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -23.16 |
| Energy contribution | -25.01 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
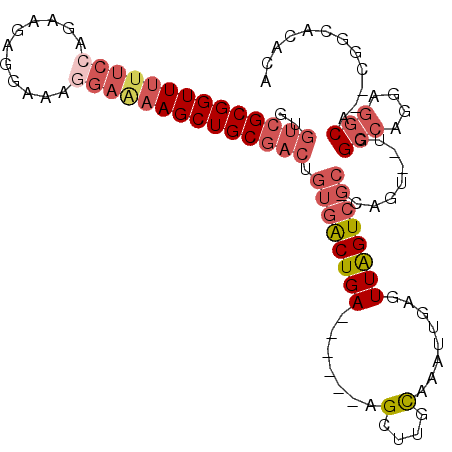
>2L_DroMel_CAF1 747022 99 - 22407834 GUGUCGCGGUUUUUCCAAAAGAGGAAACGAGAAGCUGCGACUGUGACUGA------AGCUUGCAAAUUGAGUUAGUCGC-CAGU--UGGCCAGGAGGCA--CGGCACAUA ((((((((((((((((....).(....)))))))))))((((((((((((------..(.........)..))))))).-))))--).(((....))).--.)))))... ( -37.40) >DroPse_CAF1 9921 88 - 1 AUGCCGCGGUU-UACCGCCACAG---------AGCUGC------GGCUGA------AGCUUGUAAAUUGAUUUGGUGGCACAGCUCUGGCGAGGCGACAACCGCGACAGU .((.(((((((-...((((.(((---------((((((------.((..(------(.(.........).))..)).))..)))))))....))))..))))))).)).. ( -37.50) >DroSec_CAF1 8555 99 - 1 GUGUCGCGGUUUUUCCAGAAGAGGAAAGGAAAAGCUGCGACUGUGACUGA------AGCUUGCAAAUUGAGUUAGUCGC-CAGU--UGGCCAGGAGGCA--CGGCACUCA ..((((((((((((((...........)))))))))))))).((((((((------..(.........)..))))))))-..((--(((((....))).--))))..... ( -39.70) >DroSim_CAF1 11977 105 - 1 GUGUCGCGGUUUUUCCAGAAGAGGAAAGGAAAAGCUGCGACUGUGACUGAAGCUGAAGCUUGCAAAUUGAGUUAGUCGC-CAGU--UGGCCAGGAGGCA--CGGCACACA ((((((((((((((((...........)))))))))))(((((((((((((((....)))...........))))))).-))))--).(((....))).--.)))))... ( -39.50) >DroEre_CAF1 8873 93 - 1 GUGUCGCGGUU-UUCCAGCAGAGCAAAGGAAAAGCUGCGACUGUGACUGA------AGCUUGCAAAUUGAGUUAGUCGC-CAGA--UGGCCAGCAGGCA-------CACA (((((((((((-((((.((...))...)).))))))))).((((((((((------..(.........)..))))))((-(...--.)))..)))))))-------)... ( -31.40) >DroYak_CAF1 11269 94 - 1 GUGUCGCGGUUUUUCCAGCAGAGAAAAGGAAAAGCUGCGACUGUGACUGA------AGCCUGCAAAUUGAGUUAGUCGC-CAGA--UGGCCAGGAGGCA-------CACA ..((((((((((((((...........)))))))))))))).((((((((------..(.........)..))))))))-....--(((((....))).-------)).. ( -34.60) >consensus GUGUCGCGGUUUUUCCAGAAGAGGAAAGGAAAAGCUGCGACUGUGACUGA______AGCUUGCAAAUUGAGUUAGUCGC_CAGU__UGGCCAGGAGGCA__CGGCACACA ..((((((((((((((...........)))))))))))))).((((((((.......(....)........)))))))).........(((....)))............ (-23.16 = -25.01 + 1.85)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:30 2006