
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,298,373 – 7,298,475 |
| Length | 102 |
| Max. P | 0.859217 |

| Location | 7,298,373 – 7,298,475 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -12.09 |
| Energy contribution | -14.39 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7298373 102 + 22407834 UUGGCCACAA-AAACUGUGAA---AUGCUGUGAAAAGAACGCGCGGC-CAGCAGUAGAGGUCCGAUG--ACU-UCUCUUCGGCCAGCAAAAAGUAAUUUUUGCUAAAUCG ..((((....-..(((((...---..((((((.........))))))-..)))))(((((((....)--)))-)))....))))((((((((....))))))))...... ( -36.40) >DroSec_CAF1 91278 100 + 1 UUGGCCACAA-AAACUGUGAA---AUGCUGUGAAAAGAACGCGCGGC-CAGCAGUAGAGGUCCGAAG--ACU-UC--UUUGGUCAGCAAAAAGUAAUUUUUGCUAAAUCG ..(((((...-..(((((...---..((((((.........))))))-..)))))(((((((....)--)))-))--).)))))((((((((....))))))))...... ( -34.20) >DroSim_CAF1 98091 101 + 1 UUGGCCACAA-AAACUGUGAA---AUGCUGUGAAAAGAACGCGCGGCCCAGCAGUAGAGGUCCGAAG--ACU-UC--UUUGGCCAGCAAAAAGUAAUUUUUGCUAAAUCG ..(((((...-..(((((...---..((((((.........))))))...)))))(((((((....)--)))-))--).)))))((((((((....))))))))...... ( -37.30) >DroEre_CAF1 93057 98 + 1 UUGGCCACAA-AAACUGUGAA---AUGCUGUGAAAAGAACGCGCGGC-CAGCAGCAGAGGUCCAAAA--G---UC--UGCGGCCAGCAAAAAGUAAUUUUUGCUAAAUCG ..((((....-....(((...---..((((((.........))))))-..)))(((((..(....).--.---))--)))))))((((((((....))))))))...... ( -30.90) >DroAna_CAF1 96926 97 + 1 AUGGCAGCAAAAAACUGUGCAGCAUUGCUGUGAAAAGAACGCACCAACCA-----AGAGGACCAAAA--ACU-UU--CUUGG---GCAAAAAGUAAUUUUUGCUAAAUCG .....(((((((((((.(((....(((.((((.......)))).)))(((-----(((((.......--..)-))--)))))---)))...)))..))))))))...... ( -24.10) >DroPer_CAF1 118687 98 + 1 UUGGAAGCAA-AAACUGUGAA---AUGCUGUGCAAAGAACGC-------AGCAGCAGAAGACCAGAAAGACAGUC-CAGUCUCAGACAAAAAGUAAUUUUUGCUAAAUCG .....(((((-(((((((...---.(((((((.......)))-------))))))))..........((((....-..))))..............))))))))...... ( -22.60) >consensus UUGGCCACAA_AAACUGUGAA___AUGCUGUGAAAAGAACGCGCGGC_CAGCAGUAGAGGUCCAAAA__ACU_UC__UUUGGCCAGCAAAAAGUAAUUUUUGCUAAAUCG ((((.((((......))))......((((((...................)))))).....))))...................((((((((....))))))))...... (-12.09 = -14.39 + 2.31)

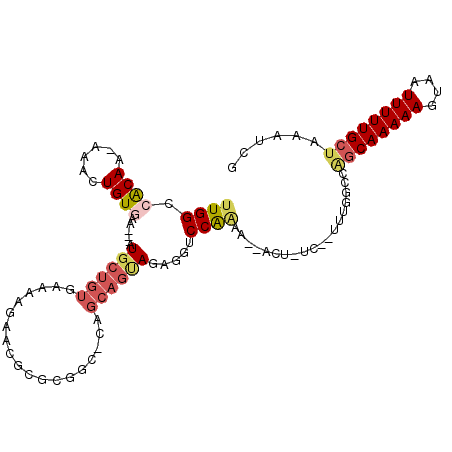
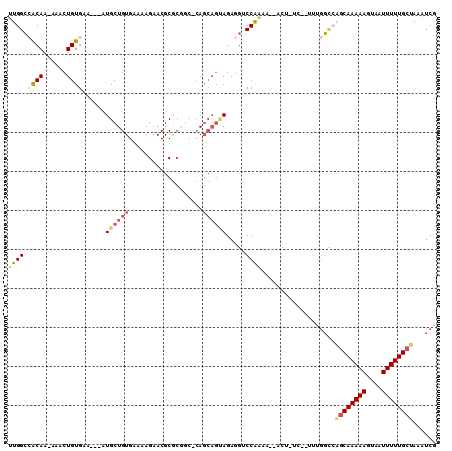
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:11 2006