
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 743,538 – 743,638 |
| Length | 100 |
| Max. P | 0.898628 |

| Location | 743,538 – 743,638 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 743538 100 - 22407834 UU----GUGGUA-UCCGCAGGUAGAAGA-CA---AUCCUCUACA-ACGGCUACAGCAGUGCAACGCAUUUUGAAACUGUUGAUGCAACGGCAGCAGAAAUUGCUGCGGAA ..----......-((((((((((((.(.-..---..).))))).-...(((.(((((((.(((......)))..)))))))..((....)))))........))))))). ( -30.80) >DroVir_CAF1 8187 98 - 1 ----------CAGCCGGCAGUUUGAAGG-UAAAAAACCACUAAA-ACGGCUACAGCGGUGCAACGCAUACUGCAACUGUUGAUGCAACGGCAGCUCAAGCUGCUGCGAAA ----------.(((((....((((..((-(.....)))..))))-.))))).((((((((((........))).)))))))......(((((((....)))))))..... ( -33.90) >DroGri_CAF1 6523 95 - 1 ----------CAUUCGACAGGUUGAAGG-CA---AACCACUACA-ACGGCUACAGCUGUGCAACGCAUUCUGCAACUGUUGAUGCAACAGCAGCUCAAAUUGCUGCGAAA ----------...(((((((((((..((-..---..))......-(((((....))))).))))((.....))..))))))).......(((((.......))))).... ( -29.60) >DroMoj_CAF1 7476 96 - 1 ----------UAACCGGCAGGUUGAAGG-CA---AACCACUAAACACGGCUACAGCGGUGCAACGCAUUCAGCAACUGUUGAUGCAACAGCAGCUCAAACUGCUGCGAAA ----------...(((((((.((((..(-(.---..........(((.((....)).)))....((((.((((....))))))))....))...)))).)))))).)... ( -30.00) >DroAna_CAF1 7822 103 - 1 UCACCCGUUG-A-ACUACAGGUAGACAG-CA---AUCCACUACA-ACGGCUACAGCGGUGCAGCGCAUUCUGAAACUGUUGAUGCAACGGCAGCAGAAGCUGCUGCGAAA ....((((((-.-.(((....)))..((-(.---..........-...))).))))))((((((((.(((((...((((((...))))))...))))))).))))))... ( -33.04) >DroPer_CAF1 8681 97 - 1 CU----CUGGU-----GCAGGUAGAAAAGCA---AUCCACUACA-ACGGCUACAGCGGUGCAACGCAUUCUGAAACUGUUGAUGCAACGCCAGCAGAAACUGCUGCGCCA ..----..(((-----((((.(((.......---........((-((((...(((.(((((...))))))))...)))))).(((.......)))....)))))))))). ( -28.90) >consensus __________UA_CCGGCAGGUAGAAGG_CA___AACCACUACA_ACGGCUACAGCGGUGCAACGCAUUCUGAAACUGUUGAUGCAACGGCAGCACAAACUGCUGCGAAA ..............................................((((..(((((((...............)))))))..))..((((((......))))))))... (-16.25 = -16.03 + -0.22)

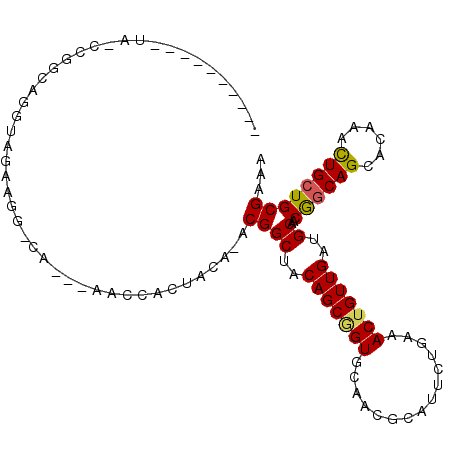
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:29 2006