| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,270,351 – 7,270,456 |
| Length | 105 |
| Max. P | 0.974655 |

| Location | 7,270,351 – 7,270,456 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
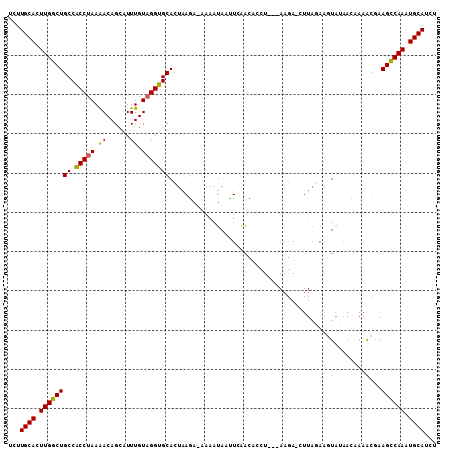
>2L_DroMel_CAF1 7270351 105 + 22407834 UCUUGCACUUGGCUGCCACCUAAAACAGCAUUUGUAGGUGCACUAAAAAAAAAUAAUUGAACAUCU---AAGA-CUUAGAAGUAUAAUAAAACGAAGUCAAAUGCAUUG ...((((.((((((((.(((((.((......)).)))))))..............((((.((.(((---(...-..)))).)).)))).......)))))).))))... ( -20.90) >DroSec_CAF1 63494 105 + 1 UCUUGCACUUGGCUGCCACCUAAGACAGCAUUUGUAGGUGCACUAAGA-AAAAUAUUUCAACUCAA---AAGAAUUUAGAAUUAUAAUAAAACGAAGCCAAAUGCAUCU ...((((.((((((((.(((((.((......)).))))))).((((((-((....))))...((..---..))..))))................)))))).))))... ( -21.90) >DroSim_CAF1 70099 105 + 1 UCUUGCACUUGGCUGCCACCUAAGACAGCAUUUGUAGGUGCACUAAGA-AAAAUAUUUCAACUGUU---AAGAAUUUAGAAUUAUCACAAAACGAAGCCAAAUGCAUCU ...((((.((((((((.(((((.((......)).))))))).((((((-((....))))..((...---.))...))))................)))))).))))... ( -22.70) >DroEre_CAF1 64415 107 + 1 UCUUGCACUUGGCUGCCGCCUAAAACAGCAUUUGUCGGUGCACUAAGA-GAAAUAGCUGGGCACCUCAGAAGA-CUUAUAAGUACAACCAAGCAAAGCCAAUUGCAUCU ...((((.((((((...((...........((((..(((((.(((..(-....)...)))))))).))))..(-((....)))........))..)))))).))))... ( -26.90) >DroYak_CAF1 64371 90 + 1 UCUUGCACUUGGCUGCCACCUAAAACAGCGUUUGUAGGUGCACUCAGA-AAAAUAACUAAGCAC---------------AAGUGAAGC---ACAAAGCCAAAUGCAACU ..(((((.((((((((.(((((((((...)))).))))))).......-...............---------------..(((...)---))..)))))).))))).. ( -22.80) >consensus UCUUGCACUUGGCUGCCACCUAAAACAGCAUUUGUAGGUGCACUAAGA_AAAAUAAUUCAACACCU___AAGA_CUUAGAAGUAUAACAAAACGAAGCCAAAUGCAUCU ...((((.((((((((.(((((.((......)).)))))))........................................((........))..)))))).))))... (-16.34 = -16.22 + -0.12)


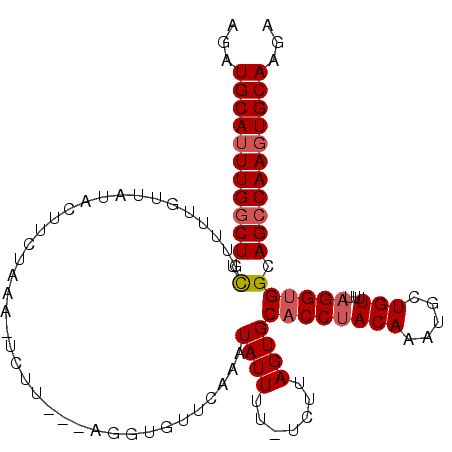
| Location | 7,270,351 – 7,270,456 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
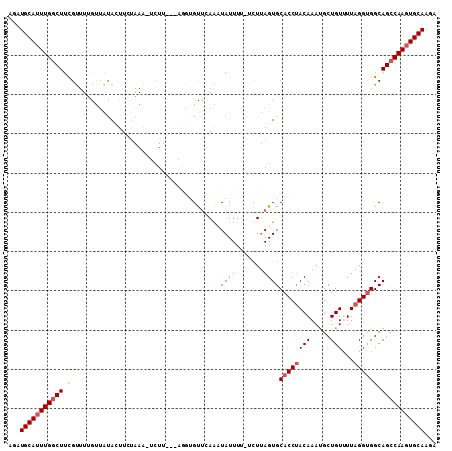
>2L_DroMel_CAF1 7270351 105 - 22407834 CAAUGCAUUUGACUUCGUUUUAUUAUACUUCUAAG-UCUU---AGAUGUUCAAUUAUUUUUUUUUAGUGCACCUACAAAUGCUGUUUUAGGUGGCAGCCAAGUGCAAGA ...((((((((.((.(..............(((((-....---(((((......)))))...)))))..((((((.((((...))))))))))).)).))))))))... ( -20.10) >DroSec_CAF1 63494 105 - 1 AGAUGCAUUUGGCUUCGUUUUAUUAUAAUUCUAAAUUCUU---UUGAGUUGAAAUAUUUU-UCUUAGUGCACCUACAAAUGCUGUCUUAGGUGGCAGCCAAGUGCAAGA ...(((((((((((.(....((((.((((((.(((....)---)))))))).))))....-........((((((((.....)))...)))))).)))))))))))... ( -28.00) >DroSim_CAF1 70099 105 - 1 AGAUGCAUUUGGCUUCGUUUUGUGAUAAUUCUAAAUUCUU---AACAGUUGAAAUAUUUU-UCUUAGUGCACCUACAAAUGCUGUCUUAGGUGGCAGCCAAGUGCAAGA ...(((((((((((.(........................---..((.(.((((....))-))..).))((((((((.....)))...)))))).)))))))))))... ( -25.30) >DroEre_CAF1 64415 107 - 1 AGAUGCAAUUGGCUUUGCUUGGUUGUACUUAUAAG-UCUUCUGAGGUGCCCAGCUAUUUC-UCUUAGUGCACCGACAAAUGCUGUUUUAGGCGGCAGCCAAGUGCAAGA ...((((.(((((((((.(((((.(((((......-....(((.(....)))).......-....))))))))))))))((((((.....))))))))))).))))... ( -34.10) >DroYak_CAF1 64371 90 - 1 AGUUGCAUUUGGCUUUGU---GCUUCACUU---------------GUGCUUAGUUAUUUU-UCUGAGUGCACCUACAAACGCUGUUUUAGGUGGCAGCCAAGUGCAAGA ..((((((((((((....---(((......---------------(..(((((.......-.)))))..)(((((.((((...)))))))))))))))))))))))).. ( -33.20) >consensus AGAUGCAUUUGGCUUCGUUUUGUUAUACUUCUAAA_UCUU___AGGUGUUCAAAUAUUUU_UCUUAGUGCACCUACAAAUGCUGUUUUAGGUGGCAGCCAAGUGCAAGA ...(((((((((((.(......................................((((.......))))((((((((.....)))...)))))).)))))))))))... (-17.98 = -18.54 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:06 2006