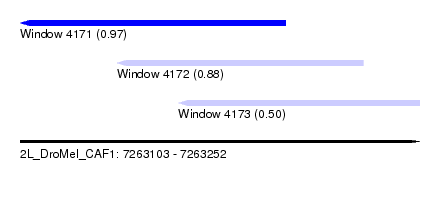
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,263,103 – 7,263,252 |
| Length | 149 |
| Max. P | 0.969525 |

| Location | 7,263,103 – 7,263,202 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 92.76 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
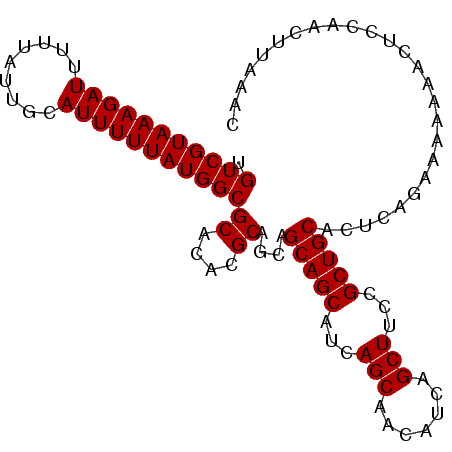
>2L_DroMel_CAF1 7263103 99 - 22407834 UGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCAGCAGCAUCAGCAACAUCAGCUUCCGCUGCACUCAGAAAAAAACUCCAACUUAAAC .(((((((((((.........)))))))))))((....))....(((((...(((.......)))...))))).......................... ( -20.90) >DroSec_CAF1 56363 99 - 1 UGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCAGCAGCAUCAGCAACAUCAGCUUCCGCUGCACUCAGAAAAAAACUCCAAUUUAAAC .(((((((((((.........)))))))))))((....))....(((((...(((.......)))...))))).......................... ( -20.90) >DroSim_CAF1 62929 99 - 1 UGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCAGCAGCAUCAGCAACAUCAGCUUCCGCUGCACUCAAAAAAAAACUCCAAUUUAAAC .(((((((((((.........)))))))))))((....))....(((((...(((.......)))...))))).......................... ( -20.90) >DroYak_CAF1 56836 92 - 1 UGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCAGCAGCAUCAGCAACAUCAGCUUCCGCUGCACUCAGAUCAAA---UUCACUU---- .(((((((((((.........)))))))))))((....))....(((((...(((.......)))...)))))............---.......---- ( -20.90) >consensus UGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCAGCAGCAUCAGCAACAUCAGCUUCCGCUGCACUCAGAAAAAAACUCCAACUUAAAC .(((((((((((.........)))))))))))((....))....(((((...(((.......)))...))))).......................... (-20.90 = -20.90 + 0.00)

| Location | 7,263,139 – 7,263,231 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7263139 92 - 22407834 ---------GCCCCAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCA------GCAGCAUCAGCAACAUCAG ---------.........(((..(((((..(((...((((((((((((((.........))))))))))))))..))).((.------...))....))))).))). ( -28.20) >DroSec_CAF1 56399 92 - 1 ---------GCCCCAAUGUGAGGGUUGCAGGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCA------GCAGCAUCAGCAACAUCAG ---------.........(((..(((((..(((...((((((((((((((.........))))))))))))))..))).((.------...))....))))).))). ( -28.00) >DroSim_CAF1 62965 92 - 1 ---------GCCCCAAUGUGAGGGUUGCAAGCCAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCA------GCAGCAUCAGCAACAUCAG ---------.........(((..(((((..((....((((((((((((((.........))))))))))))))...)).((.------...))....))))).))). ( -25.40) >DroYak_CAF1 56865 92 - 1 ---------GCCCCAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCA------GCAGCAUCAGCAACAUCAG ---------.........(((..(((((..(((...((((((((((((((.........))))))))))))))..))).((.------...))....))))).))). ( -28.20) >DroPer_CAF1 77480 104 - 1 GCACGGCCCGGCCCUAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCAUUAGGAGUAGCAGCAGCAACAU--- ((((..(((((((((.....)))))))...(((...((((((((((((((.........))))))))))))))..))).......)).)).))...........--- ( -31.00) >consensus _________GCCCCAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGCAGCA______GCAGCAUCAGCAACAUCAG ...........(((.....).))(((((..(((...((((((((((((((.........))))))))))))))..))).((..........))....)))))..... (-24.92 = -25.12 + 0.20)


| Location | 7,263,162 – 7,263,252 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
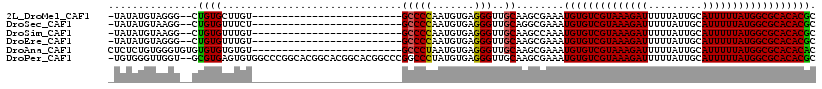
>2L_DroMel_CAF1 7263162 90 - 22407834 -UAUAUGUAGGG--CUGUGCUUGU-------------------------GCCCCAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGC -..........(--((((((((((-------------------------..(((.......)))..)))))).....(((((((((((((.........)))))))))))))))).)) ( -28.60) >DroSec_CAF1 56422 90 - 1 -UAUAUGUAAGG--CUGUGUUUCU-------------------------GCCCCAAUGUGAGGGUUGCAGGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGC -..........(--(.((((.(((-------------------------(((((.......)))..)))))......(((((((((((((.........))))))))))))))))))) ( -24.70) >DroSim_CAF1 62988 90 - 1 -UAUAUGUAAGG--CUGUGUUUGU-------------------------GCCCCAAUGUGAGGGUUGCAAGCCAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGC -.........((--((.(((....-------------------------(((((.....).)))).)))))))...((((((((((((((.........))))))))))))))..... ( -26.80) >DroEre_CAF1 57160 90 - 1 -UAUAUGUAGGG--CUGUGUUUGU-------------------------GCCCCAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGC -..........(--((((((((((-------------------------..(((.......)))..)))))).....(((((((((((((.........)))))))))))))))).)) ( -26.20) >DroAna_CAF1 57233 93 - 1 CUCUCUGUGGGUGUGUGUGUGUGU-------------------------GCCCUAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACAC ..........((((((((((.(((-------------------------(((((......)))).)))).)).......(((((((((((.........))))))))))))))))))) ( -29.00) >DroPer_CAF1 77506 115 - 1 -UGUGGGUUGGU--GCGUGAGUGUGGCCCGGCACGGCACGGCACGGCCCGGCCCUAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGC -(((((((((.(--(((((.((((......))))..))).)))))))))((((((.....))))))))).(((...((((((((((((((.........))))))))))))))..))) ( -44.30) >consensus _UAUAUGUAGGG__CUGUGUUUGU_________________________GCCCCAAUGUGAGGGUUGCAAGCGAAAUGUGUCGUAAAGAUUUUUAUUGCAUUUUUAUGGCGCACACGC ...............((((..............................(((((.......)))..))........((((((((((((((.........)))))))))))))))))). (-19.45 = -19.20 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:04 2006