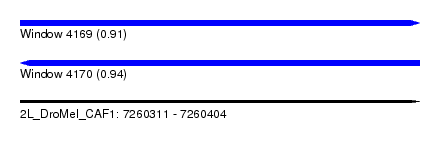
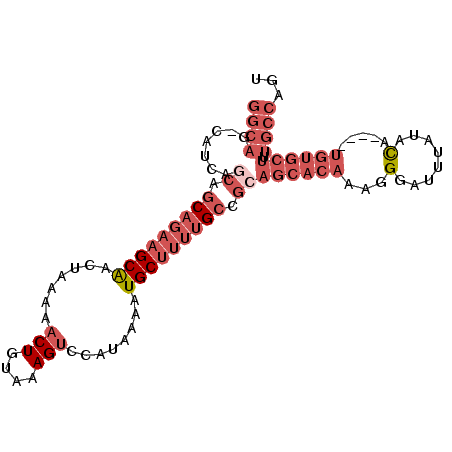
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,260,311 – 7,260,404 |
| Length | 93 |
| Max. P | 0.940559 |

| Location | 7,260,311 – 7,260,404 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -19.39 |
| Energy contribution | -20.84 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7260311 93 + 22407834 ACUGGCAAAGCACA----UGUAUAAAUCCCUUUGUGCUGCGGCAAAAGCAUUUUAUGGACUUUACAGUUUUUAGUUGCUUCUGCUGCUGAUG-CUGCC ...((((.((((((----..............))))))((((((.(((((..(((..((((....))))..))).))))).)))))).....-.)))) ( -30.24) >DroGri_CAF1 63670 84 + 1 ------------UGUGGAGAGAAAAAUCCCUUUGUGCUGUGGCAGAAGCAUUUUAUGGACU-UACAGUUU-AAGUCGCAAUAGUAGCAACAACAGCAG ------------.(..(((.((....)).)))..)(((((.((..............((((-((.....)-)))))((....)).))....))))).. ( -17.60) >DroSec_CAF1 53562 93 + 1 ACUGGCAAAGCACA----UGUAUAAAUCCCUUUGUGCUGCGGCAAAAGCAUUUUAUGGACUUUACAGGUUUUAGUUGCUUCUGCUGCUGAUG-CUGCC ...((((.((((((----..............))))))((((((.(((((......(((((.....)))))....))))).)))))).....-.)))) ( -29.24) >DroSim_CAF1 60136 93 + 1 ACUGGCAAAGCACA----UGUAUAAAUCCCUUUGUGCUGCGGCAAAAGCAUUUUAUGGACUUUACAGUUUUUAGUUGCUUCUGCUGCUGAUG-CUGCC ...((((.((((((----..............))))))((((((.(((((..(((..((((....))))..))).))))).)))))).....-.)))) ( -30.24) >DroEre_CAF1 54149 93 + 1 ACUGGCACAGCACA----UGUAUAAAUCCCUUUGUGCUGCGGCAAAAGCAUUUUACGGACUUUACAGUUUCUAGCUGCUUCUGCUGUGGAUG-CUGCC ...(((((((((((----..............)))))))(((((.(((((..(((..((((....))))..))).))))).)))))......-.)))) ( -27.64) >DroYak_CAF1 53778 93 + 1 ACGGGCAAAGCACA----UGUAUAAAUCCCUUUGUGCUGCGGCAAAAGCAUUUUAUGGACUUUACAGUUUUUAGUUGCUUCUGCCGUUGAUG-CUGCC ...((((.((((((----..............))))))((((((.(((((..(((..((((....))))..))).))))).)))))).....-.)))) ( -30.14) >consensus ACUGGCAAAGCACA____UGUAUAAAUCCCUUUGUGCUGCGGCAAAAGCAUUUUAUGGACUUUACAGUUUUUAGUUGCUUCUGCUGCUGAUG_CUGCC ...((((.((((((..................))))))((((((.(((((......(((((....))))).....))))).)))))).......)))) (-19.39 = -20.84 + 1.45)

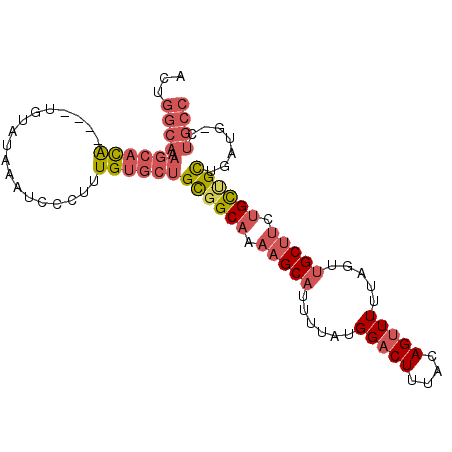

| Location | 7,260,311 – 7,260,404 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -16.15 |
| Energy contribution | -19.04 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7260311 93 - 22407834 GGCAG-CAUCAGCAGCAGAAGCAACUAAAAACUGUAAAGUCCAUAAAAUGCUUUUGCCGCAGCACAAAGGGAUUUAUACA----UGUGCUUUGCCAGU (((((-.....((.(((((((((.......(((....)))........))))))))).))((((((...(........).----)))))))))))... ( -29.06) >DroGri_CAF1 63670 84 - 1 CUGCUGUUGUUGCUACUAUUGCGACUU-AAACUGUA-AGUCCAUAAAAUGCUUCUGCCACAGCACAAAGGGAUUUUUCUCUCCACA------------ .((((((.((.((......((.(((((-(.....))-))))))......))....)).))))))...(((((....))))).....------------ ( -20.90) >DroSec_CAF1 53562 93 - 1 GGCAG-CAUCAGCAGCAGAAGCAACUAAAACCUGUAAAGUCCAUAAAAUGCUUUUGCCGCAGCACAAAGGGAUUUAUACA----UGUGCUUUGCCAGU (((((-.....((.(((((((((........((....)).........))))))))).))((((((...(........).----)))))))))))... ( -27.43) >DroSim_CAF1 60136 93 - 1 GGCAG-CAUCAGCAGCAGAAGCAACUAAAAACUGUAAAGUCCAUAAAAUGCUUUUGCCGCAGCACAAAGGGAUUUAUACA----UGUGCUUUGCCAGU (((((-.....((.(((((((((.......(((....)))........))))))))).))((((((...(........).----)))))))))))... ( -29.06) >DroEre_CAF1 54149 93 - 1 GGCAG-CAUCCACAGCAGAAGCAGCUAGAAACUGUAAAGUCCGUAAAAUGCUUUUGCCGCAGCACAAAGGGAUUUAUACA----UGUGCUGUGCCAGU (((..-........(((((((((.......(((....)))........))))))))).((((((((...(........).----)))))))))))... ( -30.26) >DroYak_CAF1 53778 93 - 1 GGCAG-CAUCAACGGCAGAAGCAACUAAAAACUGUAAAGUCCAUAAAAUGCUUUUGCCGCAGCACAAAGGGAUUUAUACA----UGUGCUUUGCCCGU (((((-......(((((((((((.......(((....)))........))))))))))).((((((...(........).----)))))))))))... ( -29.86) >consensus GGCAG_CAUCAGCAGCAGAAGCAACUAAAAACUGUAAAGUCCAUAAAAUGCUUUUGCCGCAGCACAAAGGGAUUUAUACA____UGUGCUUUGCCAGU ((((.......((.(((((((((.......(((....)))........))))))))).))((((((...(........).....)))))).))))... (-16.15 = -19.04 + 2.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:02 2006