
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,255,057 – 7,255,152 |
| Length | 95 |
| Max. P | 0.892235 |

| Location | 7,255,057 – 7,255,152 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7255057 95 - 22407834 CGGUCGGUUACAUUGAUCGCCAGUAGAUAUCAGUGUCAGU------------GGCAGUGGGCCGCCACUCGAGGCCGUUUC-AACAGCUUCAAACCCGACCAAACCGA .((((((..((((((((..(.....)..)))))))).(((------------(((.(....).)))))).(((((.((...-.)).)))))....))))))....... ( -33.40) >DroSec_CAF1 48102 95 - 1 CGGUCGGUUACAUUGAUCGCCAGUAGAUAUCAGUGUCAGU------------GGCAGUGGGCCGCCACUCGAGGCCGUUUC-AACAGCUUCAAACUCGACCAAACCGA .((((((((((((((((..(.....)..)))))))).(((------------(((.(....).)))))).(((((.((...-.)).))))).))).)))))....... ( -32.30) >DroSim_CAF1 54712 95 - 1 CGGUCGGUUACAUUGAUCGCCAGUAGAUAUCAGUGUCAGU------------GGCAGUGGGCCGCCACUCGAGGCCGUUUC-AACAGCUUCAAACUCGACCAAACCGA .((((((((((((((((..(.....)..)))))))).(((------------(((.(....).)))))).(((((.((...-.)).))))).))).)))))....... ( -32.30) >DroYak_CAF1 49138 104 - 1 CAGUCGGUUACAUUGAUCGCAAGUAGAUAUCAGUGGCAGUGGCAGUGACACUGGCAGUGGG---ACACUUGAGGCCGUUUC-AACAGCCUCAAACCUGACCAAACCGC ....(((((.((((((((....).....))))))).((.((.(((.....))).)).))((---.((.(((((((.((...-.)).)))))))...)).)).))))). ( -33.00) >DroAna_CAF1 49571 82 - 1 CGGUCGGUUACAUUGAUGACCA----AUAUCAGUGUCAGU------------GUC---------GCACCUGAGGCCGUUGA-AACAGCUUCAAACCUGACCGAACUGA ((((((((((((((((((....----.)))))))))....------------...---------.....((((((.((...-.)).)))))))))).)))))...... ( -26.70) >DroPer_CAF1 55576 86 - 1 CGGAGGGUUACAUUGAUCGC----ACACAUCAGUGUCAA---------------CAGUGU-C--CGCCCUGAGGCAGACUCAAACAGCCUCAAACCCGACAGAACCGA ((((((((.((((((...((----((......))))...---------------))))))-.--.)))))(((((...........))))).............))). ( -28.30) >consensus CGGUCGGUUACAUUGAUCGCCAGUAGAUAUCAGUGUCAGU____________GGCAGUGGGC__CCACUCGAGGCCGUUUC_AACAGCUUCAAACCCGACCAAACCGA .((((((..((((((((...........))))))))..................................(((((.((.....)).)))))....))))))....... (-18.85 = -19.18 + 0.33)

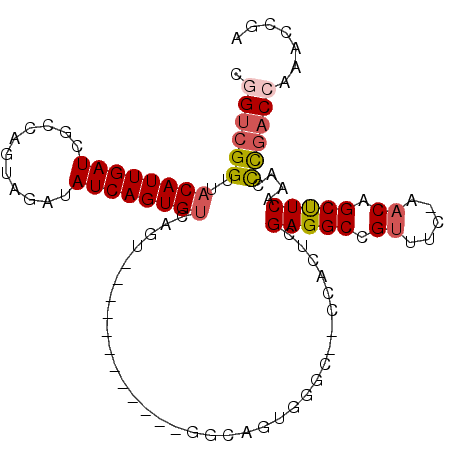
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:00 2006