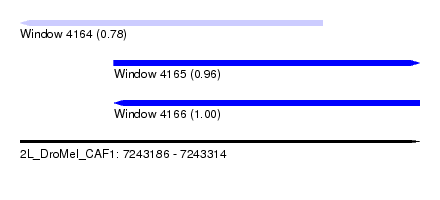
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,243,186 – 7,243,314 |
| Length | 128 |
| Max. P | 0.999954 |

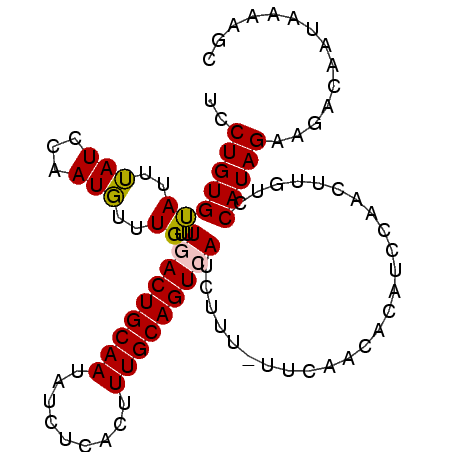
| Location | 7,243,186 – 7,243,283 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7243186 97 - 22407834 UCCUGUGUAUUUAUCCAAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCAUCUUU-UUCAACACAUCCAACUUGUCCAUAGAAGACAAUAAAAGC ..(((((.((.......(((((((..(((((((((.........))))))))).....-..))).)))).......)).))))).............. ( -18.04) >DroSec_CAF1 36380 97 - 1 UCCUGUGUAUUUAUCCAAUGUUUGUUUAACUGCAAUAUCUCACUUUGCAGUCAUCUUU-UUCAACACAUCCAACUUGUCCAUAGAAGACAAUAAAAGC ...(((((...((..((.....))..))(((((((.........))))))).......-....)))))......(((((.......)))))....... ( -13.80) >DroSim_CAF1 37955 97 - 1 UCCUGUGUAUUUAUCCAAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCAUCUUU-UUCAACACAUCCAACUUGUCCAUAGAAUACAAUAAAGGC .(((.(((((((..............(((((((((.........))))))))).....-......(((.......))).....)))))))....))). ( -19.90) >DroEre_CAF1 36985 98 - 1 GCCUGUGCAUUAAUCCAAUUAUUGUUUGACUGCAAUAUCUCACUUUGCAGUGAGAUUUAUUCAACACCUCCAGCUUGUCCAUAGAUGGCAAAAAAUGC (((...((...........((((((......))))))(((((((....))))))).................))..(((....))))))......... ( -18.30) >DroYak_CAF1 36706 88 - 1 UCCUGUGCAUUUAUGCCAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCA----------UCUCAUCCCACACAUCCAUAGAUGACAAUAAAAGC ...(((.((((((((...(((.((..(((((((((.........)))))))))----------...))....)))....)))))))))))........ ( -22.30) >consensus UCCUGUGUAUUUAUCCAAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCAUCUUU_UUCAACACAUCCAACUUGUCCAUAGAAGACAAUAAAAGC ..(((((((..(((...)))..))..(((((((((.........)))))))))..........................))))).............. (-11.74 = -11.58 + -0.16)

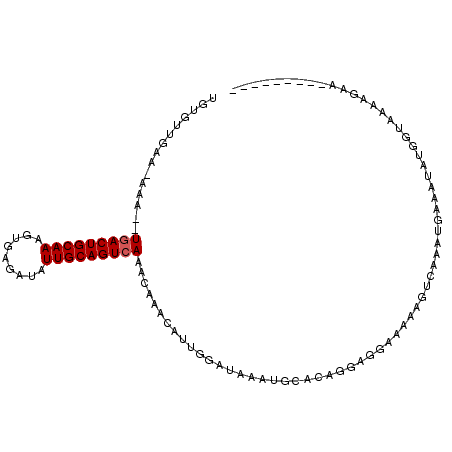
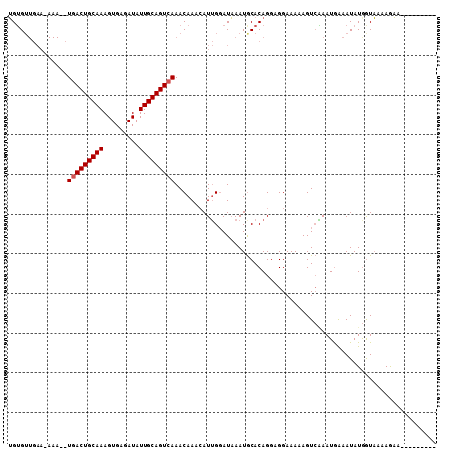
| Location | 7,243,216 – 7,243,314 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -16.63 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7243216 98 + 22407834 UGUGUUGAA-AAAGAUGACUGCAAAGUGAGAUAUUGCAGUCAAACAAACAUUGGAUAAAUACACAGGAGGAAAAAGUGGAAUGGAAUAUAAUAAAAGAA--------- ((((((...-.....(((((((((.........)))))))))...................(((...........)))......)))))).........--------- ( -14.80) >DroEre_CAF1 37015 108 + 1 GGUGUUGAAUAAAUCUCACUGCAAAGUGAGAUAUUGCAGUCAAACAAUAAUUGGAUUAAUGCACAGGCGGAAAAAGUCAAAUGAAAUAUGGUCAAAACAAGUAUUCCA .((.((((....((((((((....))))))))..((((.((((.......)))).....))))..(((.......))).............)))).)).......... ( -19.50) >DroYak_CAF1 36736 89 + 1 UGAGA----------UGACUGCAAAGUGAGAUAUUGCAGUCAAACAAACAUGGCAUAAAUGCACAGGAGGAAAAAGUCUAAUAAAAUAUGGUAAAAGAA--------- ..(((----------(((((((((.........)))))))).........(((((....))).))..........))))....................--------- ( -15.60) >consensus UGUGUUGAA_AAA__UGACUGCAAAGUGAGAUAUUGCAGUCAAACAAACAUUGGAUAAAUGCACAGGAGGAAAAAGUCAAAUGAAAUAUGGUAAAAGAA_________ ...............(((((((((.........))))))))).................................................................. ( -9.80 = -10.13 + 0.33)

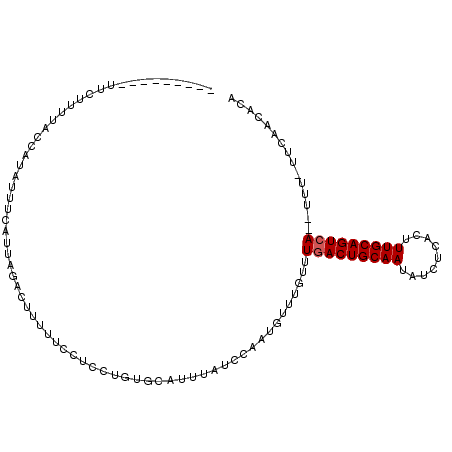
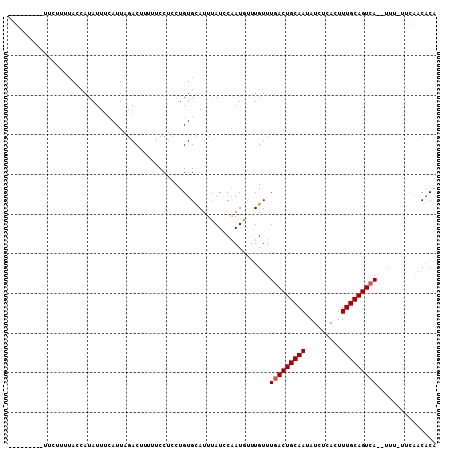
| Location | 7,243,216 – 7,243,314 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -15.63 |
| Consensus MFE | -10.33 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.83 |
| SVM RNA-class probability | 0.999954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7243216 98 - 22407834 ---------UUCUUUUAUUAUAUUCCAUUCCACUUUUUCCUCCUGUGUAUUUAUCCAAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCAUCUUU-UUCAACACA ---------...................................((((.......((.....))..(((((((((.........))))))))).....-....)))). ( -14.10) >DroEre_CAF1 37015 108 - 1 UGGAAUACUUGUUUUGACCAUAUUUCAUUUGACUUUUUCCGCCUGUGCAUUAAUCCAAUUAUUGUUUGACUGCAAUAUCUCACUUUGCAGUGAGAUUUAUUCAACACC .((((.....(((.(((.......)))...)))...))))....(((............((((((......))))))(((((((....))))))).........))). ( -18.50) >DroYak_CAF1 36736 89 - 1 ---------UUCUUUUACCAUAUUUUAUUAGACUUUUUCCUCCUGUGCAUUUAUGCCAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCA----------UCUCA ---------..................................((.(((....)))))........(((((((((.........)))))))))----------..... ( -14.30) >consensus _________UUCUUUUACCAUAUUUCAUUAGACUUUUUCCUCCUGUGCAUUUAUCCAAUGUUUGUUUGACUGCAAUAUCUCACUUUGCAGUCA__UUU_UUCAACACA ..................................................................(((((((((.........)))))))))............... (-10.33 = -10.67 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:58 2006