
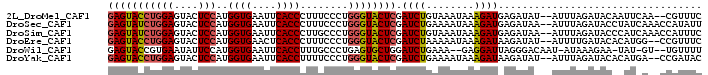
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,220,626 – 7,220,721 |
| Length | 95 |
| Max. P | 0.927944 |

| Location | 7,220,626 – 7,220,721 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7220626 95 + 22407834 GAGUACCUGGAGUACUCCAUGGUGAAUUCACCCUUUCCCUGGGUACUCGAUCUGUAAAUAAAGAUGAGAUAU--AUUUAGAUACAAUUCAA--CGUUUC (((((((((((....)))..((((....))))........))))))))((..((((..((((..........--.))))..))))..))..--...... ( -25.60) >DroSec_CAF1 14579 97 + 1 GAGUAUCUGGAGUACUCCAUGGUGAAUUCACCCUUUCCCUGGGUACUCGAUCUGAAAAUAAAGAUGAGAUAA--AUUUAGAUACCUAUCAAACCAUAUU ..(((((((((((((.(((.((.(((........)))))))))))))).((((........)))).......--...)))))))............... ( -25.70) >DroSim_CAF1 14827 97 + 1 GAGUAUCUGGAGUACUCCAUGGUGAAUUCACCCUUGCCCUGGGUACUCGAUCUGUAAAUAAAGAUGAGAUAA--AUUUAGAUACCCAUCAAACCAUUUC (.(((((((((((((.(((.((..(........)..)).))))))))).((((........)))).......--...))))))).)............. ( -27.20) >DroEre_CAF1 15181 95 + 1 GAGUACCUGGAGUACUCCAUGGUGAACUCACCCUUUCCCUGGGUACUCGAUCUAAAAAUAAAGAUAAGAUAU--AUUUUGAUACACAUGG--CCGUUUC (((((((((((....)))..((((....))))........)))))))).((((........)))).......--................--....... ( -22.10) >DroWil_CAF1 24296 92 + 1 GAGUACCGUGAAUAUUCCAUGGUGAAUUCACCUUUGCCCUGAGUGCUGGAUCUGAAA--GAGGAUUAGGGACAAU-AUAAAGAA-UAU-GU--UGUUUU ((((((((((.......))))))..)))).......((((((...((.........)--)....))))))(((((-(((.....-)))-))--)))... ( -21.20) >DroYak_CAF1 15009 95 + 1 GAGUACCUGGAGUACUCCAUGGUGAAUUCACCUUUUCCCUGGGUACUCGAUCUGAAAAUAAAGAUAAGAUAU--AUUUAGAUACACAUGA--CCGAUAC (((((((((((....)))..((((....))))........)))))))).(((((((.(((.........)))--.)))))))........--....... ( -25.40) >consensus GAGUACCUGGAGUACUCCAUGGUGAAUUCACCCUUUCCCUGGGUACUCGAUCUGAAAAUAAAGAUGAGAUAU__AUUUAGAUACACAUCAA_CCGUUUC (((((((((((....)))..((((....))))........)))))))).((((........)))).................................. (-18.86 = -19.03 + 0.17)
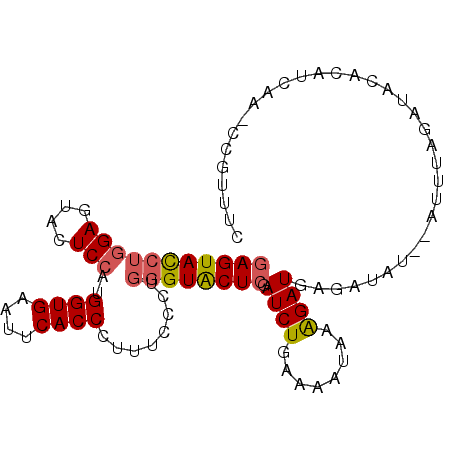

| Location | 7,220,626 – 7,220,721 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -22.11 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7220626 95 - 22407834 GAAACG--UUGAAUUGUAUCUAAAU--AUAUCUCAUCUUUAUUUACAGAUCGAGUACCCAGGGAAAGGGUGAAUUCACCAUGGAGUACUCCAGGUACUC ......--..((.(((((..((((.--((.....)).))))..))))).))(((((((.........((((....)))).((((....))))))))))) ( -24.40) >DroSec_CAF1 14579 97 - 1 AAUAUGGUUUGAUAGGUAUCUAAAU--UUAUCUCAUCUUUAUUUUCAGAUCGAGUACCCAGGGAAAGGGUGAAUUCACCAUGGAGUACUCCAGAUACUC ..............(((((((....--.......((((........)))).(((((((((.......((((....)))).))).)))))).))))))). ( -23.50) >DroSim_CAF1 14827 97 - 1 GAAAUGGUUUGAUGGGUAUCUAAAU--UUAUCUCAUCUUUAUUUACAGAUCGAGUACCCAGGGCAAGGGUGAAUUCACCAUGGAGUACUCCAGAUACUC .............((((((((....--.......((((........)))).(((((((((.......((((....)))).))).)))))).)))))))) ( -25.10) >DroEre_CAF1 15181 95 - 1 GAAACGG--CCAUGUGUAUCAAAAU--AUAUCUUAUCUUUAUUUUUAGAUCGAGUACCCAGGGAAAGGGUGAGUUCACCAUGGAGUACUCCAGGUACUC .......--......(((((.....--.......((((........)))).(((((((((.......((((....)))).))).))))))..))))).. ( -22.00) >DroWil_CAF1 24296 92 - 1 AAAACA--AC-AUA-UUCUUUAU-AUUGUCCCUAAUCCUC--UUUCAGAUCCAGCACUCAGGGCAAAGGUGAAUUCACCAUGGAAUAUUCACGGUACUC ......--..-(((-((((....-.(((.((((.......--.................))))))).((((....))))..)))))))........... ( -14.76) >DroYak_CAF1 15009 95 - 1 GUAUCGG--UCAUGUGUAUCUAAAU--AUAUCUUAUCUUUAUUUUCAGAUCGAGUACCCAGGGAAAAGGUGAAUUCACCAUGGAGUACUCCAGGUACUC (((((((--((((((((......))--))))..((....))......))))(((((((((.......((((....)))).))).))))))..))))).. ( -22.90) >consensus GAAACGG_UUGAUGUGUAUCUAAAU__AUAUCUCAUCUUUAUUUUCAGAUCGAGUACCCAGGGAAAGGGUGAAUUCACCAUGGAGUACUCCAGGUACUC ...............((((((.............((((........)))).(((((((((.......((((....)))).))).)))))).)))))).. (-15.94 = -16.63 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:53 2006