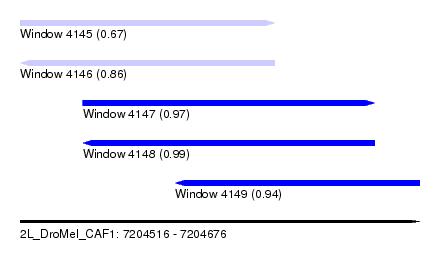
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,204,516 – 7,204,676 |
| Length | 160 |
| Max. P | 0.992155 |

| Location | 7,204,516 – 7,204,618 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
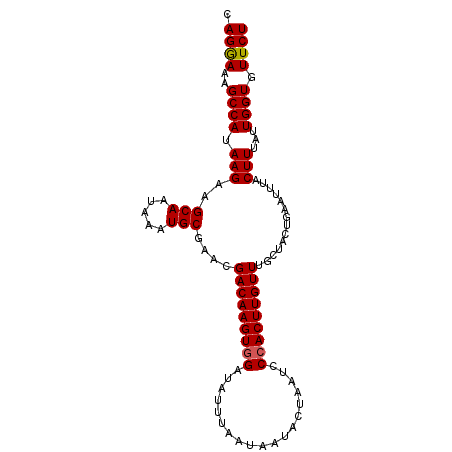
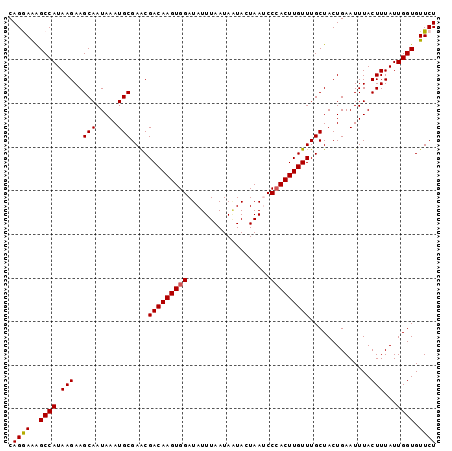
>2L_DroMel_CAF1 7204516 102 + 22407834 UAAAUGAGUUAAACACUGUUUGGUAUCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUUGGA ...........(((..((((((.(((.(...(((((((.((((((.(((((.(((....))).)))))..)))))))))))))).))).)))))).)))... ( -18.70) >DroSec_CAF1 22129 102 + 1 UUAAUAAGCUAAAUACUGUUUGGUAUCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGG .............(((((((((.(((.(...(((((((.((((((.(((((.(((....))).)))))..)))))))))))))).))).))))).))))... ( -20.20) >DroSim_CAF1 35358 102 + 1 UUAAUAAGUUAAACACUGUUUGGUAUCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGG .............(((((((((.(((.(...(((((((.((((((.(((((.(((....))).)))))..)))))))))))))).))).))))).))))... ( -22.50) >DroEre_CAF1 47443 102 + 1 UAAAAAAGUCAACCUUUGUUUGGUAUCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGA ............((((((((((.(((.(...(((((((.((((((.(((((.(((....))).)))))..)))))))))))))).))).)))))))).)).. ( -23.00) >DroYak_CAF1 37759 102 + 1 UAAAAAACUUUACCCUUAUUUGGUAUCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGA ............(((((.((((.(((.(...(((((((.((((((.(((((.(((....))).)))))..)))))))))))))).))).)))).))).)).. ( -19.70) >consensus UAAAUAAGUUAAACACUGUUUGGUAUCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGA ................((((((.(((.(...(((((((.((((((.(((((.(((....))).)))))..)))))))))))))).))).))))))....... (-17.48 = -17.68 + 0.20)

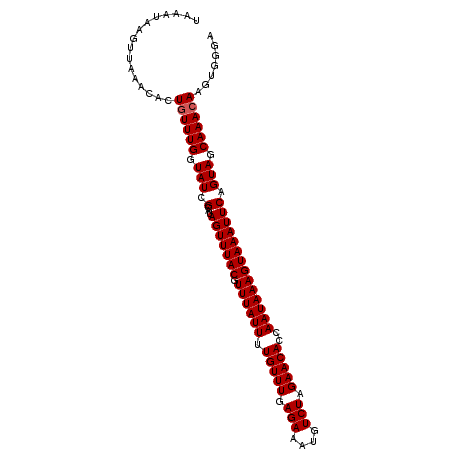
| Location | 7,204,516 – 7,204,618 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -16.56 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7204516 102 - 22407834 UCCAACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGAUACCAAACAGUGUUUAACUCAUUUA ...((((((((((.((.((..(((((((((((..((((..(((....)))..)))).)))))).))))).....)).)).))))))).)))........... ( -15.80) >DroSec_CAF1 22129 102 - 1 CCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGAUACCAAACAGUAUUUAGCUUAUUAA ....(((.(((((.((.((..(((((((((((..((((..(((....)))..)))).)))))).))))).....)).)).)))))))).............. ( -14.90) >DroSim_CAF1 35358 102 - 1 CCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGAUACCAAACAGUGUUUAACUUAUUAA ...((((.(((((.((.((..(((((((((((..((((..(((....)))..)))).)))))).))))).....)).)).)))))))))............. ( -17.40) >DroEre_CAF1 47443 102 - 1 UCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGAUACCAAACAAAGGUUGACUUUUUUA ..((..(((((((.((.((..(((((((((((..((((..(((....)))..)))).)))))).))))).....)).)).))))))).))............ ( -16.00) >DroYak_CAF1 37759 102 - 1 UCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGAUACCAAAUAAGGGUAAAGUUUUUUA ..((.((((((((.((.((..(((((((((((..((((..(((....)))..)))).)))))).))))).....)).)).))))))))))............ ( -18.70) >consensus UCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGAUACCAAACAGUGUUUAACUUAUUUA ......(((((((.((.((..(((((((((((..((((..(((....)))..)))).)))))).))))).....)).)).)))))))............... (-14.92 = -14.52 + -0.40)


| Location | 7,204,541 – 7,204,658 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 98.21 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -24.64 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7204541 117 + 22407834 UCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUUGGAUUAGUAAUAUUAAAUAUCCACUUGUCGUUCGCAUUUAUUG ..((..(((((((.((((((.(((((.(((....))).)))))..)))))))))))))..(.(((..((((((.((((...((....))...)))))))))).))))))........ ( -23.10) >DroSec_CAF1 22154 117 + 1 UCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGGUUAGUAUUAUUAAAUAUCCACUUGUCGUUCGCAUUUAUUG ..((..(((((((.((((((.(((((.(((....))).)))))..)))))))))))))..(.(((..(((((((((.(((((...)))))...))))))))).))))))........ ( -26.60) >DroSim_CAF1 35383 117 + 1 UCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGGUUAGUAUUAUUAAAUAUCCACUUGUCGUUCGCAUUUAUUG ..((..(((((((.((((((.(((((.(((....))).)))))..)))))))))))))..(.(((..(((((((((.(((((...)))))...))))))))).))))))........ ( -26.60) >DroEre_CAF1 47468 117 + 1 UCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGAUUAGUACUAUUAAAUAUCCACUUGUCGCUCGCAUUUAUUG ..((..(((((((.((((((.(((((.(((....))).)))))..)))))))))))))..(.(((..(((((((((.(((((...)))))...))))))))).))))))........ ( -26.80) >DroYak_CAF1 37784 117 + 1 UCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGAUUAGUAUUAUUAAAUAUCCACUUGUCGUUCGCAUUUAUUG ..((..(((((((.((((((.(((((.(((....))).)))))..)))))))))))))..(.(((..(((((((((.(((((...)))))...))))))))).))))))........ ( -24.40) >consensus UCGCAUAGUUUACGUUUAUUUUGUUUGAGAAAUGUCUAGAACACCAAUAAAGUAAAUUCAGUAGCAAACAAGUGGGAUUAGUAUUAUUAAAUAUCCACUUGUCGUUCGCAUUUAUUG ..((..(((((((.((((((.(((((.(((....))).)))))..)))))))))))))..(.(((..(((((((((.(((((...)))))...))))))))).))))))........ (-24.64 = -24.68 + 0.04)

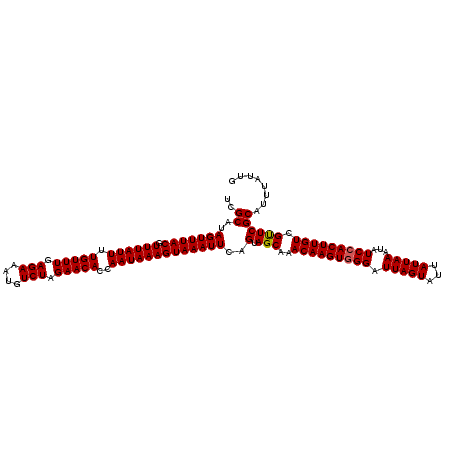
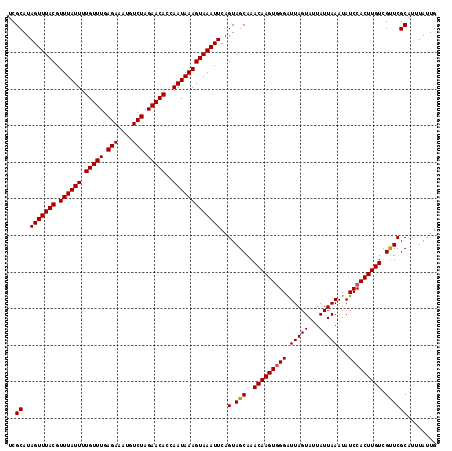
| Location | 7,204,541 – 7,204,658 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 98.21 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.53 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7204541 117 - 22407834 CAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAUUACUAAUCCAACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGA ........((((((((....(((((..............)))))..)))))))).......(((((((((((..((((..(((....)))..)))).)))))).)))))........ ( -21.74) >DroSec_CAF1 22154 117 - 1 CAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAACCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGA .......((((...(((((((((...................)))))))))..........(((((((((((..((((..(((....)))..)))).)))))).)))))...)))). ( -22.61) >DroSim_CAF1 35383 117 - 1 CAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAACCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGA .......((((...(((((((((...................)))))))))..........(((((((((((..((((..(((....)))..)))).)))))).)))))...)))). ( -22.61) >DroEre_CAF1 47468 117 - 1 CAAUAAAUGCGAGCGACAAGUGGAUAUUUAAUAGUACUAAUCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGA .......((((((((((((((((...................))))))).)))))......(((((((((((..((((..(((....)))..)))).)))))).)))))...)))). ( -26.41) >DroYak_CAF1 37784 117 - 1 CAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAAUCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGA .......((((...(((((((((...................)))))))))..........(((((((((((..((((..(((....)))..)))).)))))).)))))...)))). ( -22.61) >consensus CAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAAUCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCUAGACAUUUCUCAAACAAAAUAAACGUAAACUAUGCGA .......((((...(((((((((...................)))))))))..........(((((((((((..((((..(((....)))..)))).)))))).)))))...)))). (-21.33 = -21.53 + 0.20)

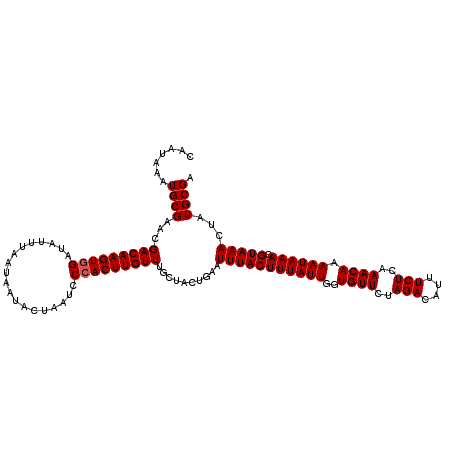
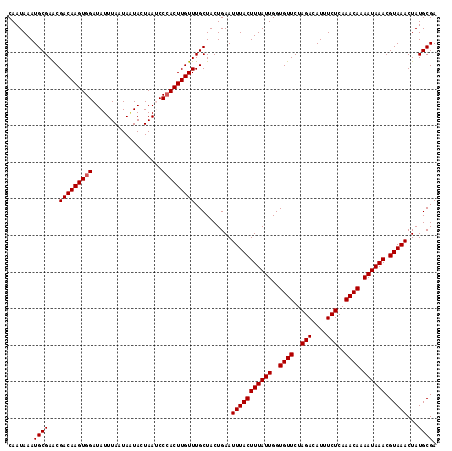
| Location | 7,204,578 – 7,204,676 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 96.43 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.69 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7204578 98 - 22407834 CAGAAGAGCCAUAAGAAGCAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAUUACUAAUCCAACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCU .((((..((((.(((..(((.....)))....(((((((((((..............)))).)))))))..............)))...)))).)))) ( -19.64) >DroSec_CAF1 22191 98 - 1 CAGGAAGGCCAUAAGAAGCAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAACCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCU ..((((.((((.(((..(((.....)))....(((((((((...................)))))))))..............)))...)))).)))) ( -23.31) >DroSim_CAF1 35420 98 - 1 CAGGAAAGCCAUAAGAAGCAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAACCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCU ..((((.((((.(((..(((.....)))....(((((((((...................)))))))))..............)))...)))).)))) ( -21.21) >DroEre_CAF1 47505 98 - 1 CAGGAAAGCCAUAAGAAGCAAUAAAUGCGAGCGACAAGUGGAUAUUUAAUAGUACUAAUCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCU ..((((.((((.(((..(((.....))).((((((((((((...................))))))).)))))..........)))...)))).)))) ( -25.01) >DroYak_CAF1 37821 98 - 1 CAGAAAAGCCAUAAGAAGCAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAAUCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCU .((((..((((.(((..(((.....)))....(((((((((...................)))))))))..............)))...)))).)))) ( -19.71) >consensus CAGGAAAGCCAUAAGAAGCAAUAAAUGCGAACGACAAGUGGAUAUUUAAUAAUACUAAUCCCACUUGUUUGCUACUGAAUUUACUUUAUUGGUGUUCU .((((..((((.(((..(((.....)))....(((((((((...................)))))))))..............)))...)))).)))) (-18.73 = -18.69 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:42 2006