| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,198,535 – 7,198,643 |
| Length | 108 |
| Max. P | 0.927917 |

| Location | 7,198,535 – 7,198,643 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.76 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -10.15 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7198535 108 + 22407834 GGAGUUU------UUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUUUUUUCGUCCUUAAGUGAAUGCAAACUCUAUAAAAAGCAGGGUGACACAAGUC-AU--- (((((((------...((.(((((((((((.(((...(((((((......)))))))....))).))...))))))))))))))))))............(((((....)))-))--- ( -22.10) >DroSec_CAF1 16145 110 + 1 GGAGUUU------UUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUUUUUU-GGCCUUAAGUGAAGGCAAACUCUAUCAAAAGCAGGGUGACACAAAUC-AUAAC (((((((------...(((.((((((((((((((...(((((((......)))))))....)))-))..)))))))))))))))))))...........(....).......-..... ( -28.00) >DroSim_CAF1 29677 91 + 1 GGAGUUU------UUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUU------------------GGCAAACU--AUAAAAAGCAGGGUGACACAAAUC-AUAAC ..(((((------...((((.((((((....))))))(((((((......)))))))..)------------------))))))))--...........(....).......-..... ( -15.40) >DroEre_CAF1 40924 104 + 1 GGAGUUU-----UUUCGUCAUUUGUUUAACCAAAUAAAUGUCAACAAUGUUUGACGUAUUUGUAGCCUCUUAA-----GGCAAACUCUAUAAAAAACAGGCUGACUCGGAUC-GG--- .(((...-----.)))(((((((((((...((((((...(((((......))))).))))))..((((....)-----)))............))))))).)))).......-..--- ( -21.70) >DroYak_CAF1 31463 109 + 1 GAAGUUU-----UUUCGCCAUUUAUUUAACCAAAUAAAUGUCAGCAAUGGCUGACGUAUUUUUACUUCCUUAAGUUUGGGCAAACUCUAUAAGCAGUAGGGUGACUCCAAAC-GU--- (((((..-----....((.((((((((....))))))))((((((....))))))))......))))).....(((((((...(((((((.....)))))))...)))))))-..--- ( -32.00) >DroAna_CAF1 17046 116 + 1 ACAUUUCCAGAGUUUCGCCAUUUAUCUAACCAAAUAAAUGUCAACAAUGGCUGACAUAUAUUCCAAUCCUCGAACGCAGACGGUCUUUUGAGAUACGAGGAUACCACCAGAUAAG-G- ....................(((((((.....((((.((((((........)))))).))))...(((((((..((....))((((....)))).)))))))......)))))))-.- ( -23.50) >consensus GGAGUUU______UUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUUUUUA_GUCCUUAAGUG_AGGCAAACUCUAUAAAAAGCAGGGUGACACAAAUC_AU___ ..............(((((..((((((....))))))(((((((......)))))))..........................................))))).............. (-10.15 = -10.57 + 0.42)

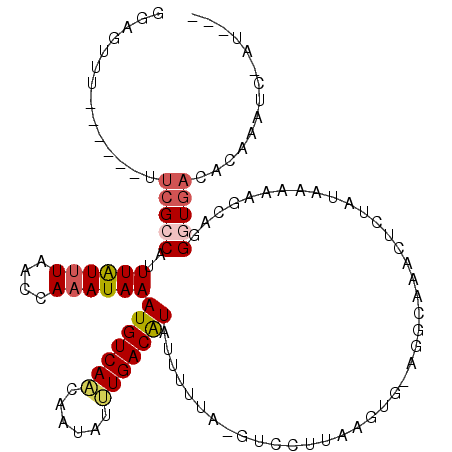

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:34 2006