
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,194,113 – 7,194,205 |
| Length | 92 |
| Max. P | 0.679317 |

| Location | 7,194,113 – 7,194,205 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -32.08 |
| Energy contribution | -33.30 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
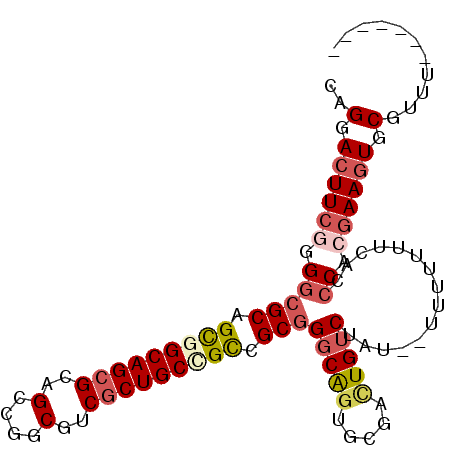
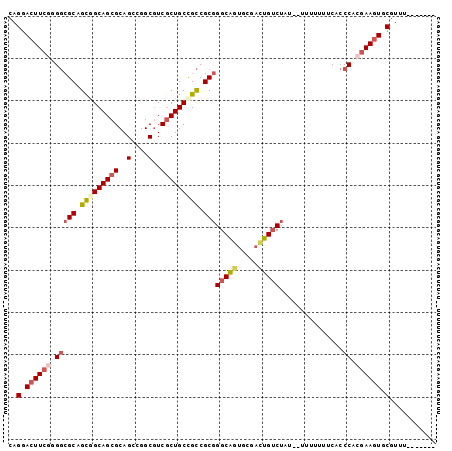
>2L_DroMel_CAF1 7194113 92 + 22407834 CAGGACUUCGGGGCGCAGCGGCAGCGCAGCCGGCGUCGCUGCCGCCGCGGGCAGUGCGACUGUCUAU--UUUUUUUCACCCACGAAGUGCGUUU------- ..(.((((((((((((.(((((((((..(....)..))))))))).)))(((((.....)))))...--.........))).)))))).)....------- ( -46.00) >DroSec_CAF1 11816 92 + 1 CAGGACUUCGAGGCGCAGCGGCAGCGCAGCCGGCAUCGCUGCCGCCGCGGGCAGUGCGACUGUCAAU--UUUUUUUCACCCACGAAGUGCGUUU------- ..(.((((((.(((((.(((((((((..(....)..))))))))).)))(((((.....)))))...--..........)).)))))).)....------- ( -42.50) >DroSim_CAF1 25338 92 + 1 CAGGACUUCAAGGCGCAGUAGCAGCGCAGCCGGCGUCGCUGCCGCCGCGGGCGGUGCGACUGUCUAU--UUUUUUUCACCCACGAAGUGCGUUU------- ..(.(((((..(((((.((....)))).)))((((((((.((((((...))))))))))).)))...--..............))))).)....------- ( -35.80) >DroEre_CAF1 34246 91 + 1 CAGGACUUCGGGGCGCAGCGGCAGCGCAGCCGGCGUCGCUGCCGCCGCGGGCAGUGCGACUGUCUAU--UUU-UUUCACCCACGAAGUGCGUUU------- ..(.((((((((((((.(((((((((..(....)..))))))))).)))(((((.....)))))...--...-.....))).)))))).)....------- ( -46.00) >DroYak_CAF1 26711 92 + 1 CAGGACUUCGGGGCGCAGCAGCAGCGCAGCCGGCGUCGCUGCCGCCGCGGGCAUUGCGACUGUCUAU--UUUUUUUCACCCACGAAGUGCGUUU------- ..(.(((((((((.((.(((((.((((.....)))).))))).))...(((((.......)))))..--.........))).)))))).)....------- ( -36.50) >DroAna_CAF1 12484 100 + 1 CAGGACUUCGUG-AGCAGCGGCAGCGCAGUAGGCGUCUCUGCUGUCGCGGGCAGAGAGGUUGACUGCUCUUUUUUUCUACCAAAAACUGCGUGCCUAUCGA ...(.((.(...-.).)))(((((((((((..((.((((((((......)))))))).))..)))))...(((((......)))))..)).))))...... ( -35.10) >consensus CAGGACUUCGGGGCGCAGCGGCAGCGCAGCCGGCGUCGCUGCCGCCGCGGGCAGUGCGACUGUCUAU__UUUUUUUCACCCACGAAGUGCGUUU_______ ..(.((((((.(((((.(((((((((..(....)..))))))))).)))(((((.....)))))...............)).)))))).)........... (-32.08 = -33.30 + 1.22)

| Location | 7,194,113 – 7,194,205 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -31.39 |
| Energy contribution | -32.37 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
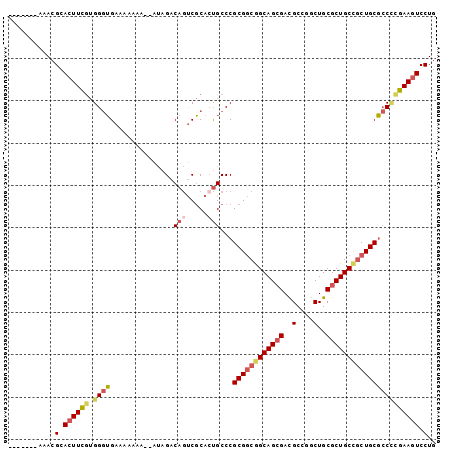
>2L_DroMel_CAF1 7194113 92 - 22407834 -------AAACGCACUUCGUGGGUGAAAAAAA--AUAGACAGUCGCACUGCCCGCGGCGGCAGCGACGCCGGCUGCGCUGCCGCUGCGCCCCGAAGUCCUG -------....(.((((((.((((........--.....(((.....)))...((((((((((((..(....)..)))))))))))))))))))))).).. ( -44.00) >DroSec_CAF1 11816 92 - 1 -------AAACGCACUUCGUGGGUGAAAAAAA--AUUGACAGUCGCACUGCCCGCGGCGGCAGCGAUGCCGGCUGCGCUGCCGCUGCGCCUCGAAGUCCUG -------....(.((((((.((((((......--........))))......(((((((((((((..(....)..))))))))))))))).)))))).).. ( -40.24) >DroSim_CAF1 25338 92 - 1 -------AAACGCACUUCGUGGGUGAAAAAAA--AUAGACAGUCGCACCGCCCGCGGCGGCAGCGACGCCGGCUGCGCUGCUACUGCGCCUUGAAGUCCUG -------....(.(((((((((((((......--........)))).)))).(((((..((((((..(....)..))))))..)))))....))))).).. ( -33.94) >DroEre_CAF1 34246 91 - 1 -------AAACGCACUUCGUGGGUGAAA-AAA--AUAGACAGUCGCACUGCCCGCGGCGGCAGCGACGCCGGCUGCGCUGCCGCUGCGCCCCGAAGUCCUG -------....(.((((((.((((....-...--.....(((.....)))...((((((((((((..(....)..)))))))))))))))))))))).).. ( -44.00) >DroYak_CAF1 26711 92 - 1 -------AAACGCACUUCGUGGGUGAAAAAAA--AUAGACAGUCGCAAUGCCCGCGGCGGCAGCGACGCCGGCUGCGCUGCUGCUGCGCCCCGAAGUCCUG -------....(.((((((.(((((.......--.......(((((.......)))))(((((((.(((.....))).))))))).))))))))))).).. ( -39.30) >DroAna_CAF1 12484 100 - 1 UCGAUAGGCACGCAGUUUUUGGUAGAAAAAAAGAGCAGUCAACCUCUCUGCCCGCGACAGCAGAGACGCCUACUGCGCUGCCGCUGCU-CACGAAGUCCUG ....((((.(((((((....(((((.........(((((...(.(((((((........))))))).)...))))).)))))))))).-......)))))) ( -29.91) >consensus _______AAACGCACUUCGUGGGUGAAAAAAA__AUAGACAGUCGCACUGCCCGCGGCGGCAGCGACGCCGGCUGCGCUGCCGCUGCGCCCCGAAGUCCUG ...........(.((((((.((((...............(((.....)))...((((((((((((..(....)..)))))))))))))))))))))).).. (-31.39 = -32.37 + 0.98)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:32 2006