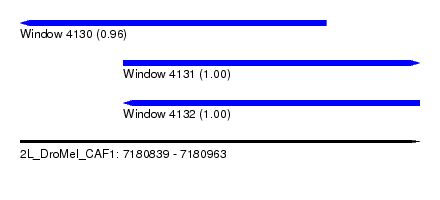
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,180,839 – 7,180,963 |
| Length | 124 |
| Max. P | 0.999895 |

| Location | 7,180,839 – 7,180,934 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -21.32 |
| Energy contribution | -22.33 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7180839 95 - 22407834 GAUUGGCCUUUGGGGGCAUCCCAAUAAUGGAGCCAGACGUGGCUAACUUUUAUCAGUUAGCCG------GGCUGUAGGGC-CGGCUAAAUCAAUGCAAUUAA (((((.((.(((((.....)))))....))((((.....(((((((((......)))))))))------((((....)))-))))).........))))).. ( -33.60) >DroSec_CAF1 122242 95 - 1 GAUUGGCCUUUGGGGGCAACCCAAUAAUGGAGCCAGACGUGGCCAACUUUUAUUAGUUAGCCG------GGCUGUAGGGC-CUGCUAAAUCAAUGCAAUUAA ....(((((...(((....))).....((.((((.....((((.((((......)))).))))------)))).))))))-)(((.........)))..... ( -31.60) >DroAna_CAF1 130679 94 - 1 UAUUGGCA-------AGUACUCUAUAAUGGAGAUGGAUA-GGCCAACUUUUAUCGGUUGGUUGGCUGUAGGCUGUAGGGCCCUGCUAAAUCAAUGCAAUUAA .....(((-------....((((.....))))...(((.-((((((((..........))))))))(((((((....)).)))))...)))..)))...... ( -24.80) >consensus GAUUGGCCUUUGGGGGCAACCCAAUAAUGGAGCCAGACGUGGCCAACUUUUAUCAGUUAGCCG______GGCUGUAGGGC_CUGCUAAAUCAAUGCAAUUAA ..((((((....(((....))).....((.((((.....(((((((((......)))))))))......)))).))......)))))).............. (-21.32 = -22.33 + 1.01)



| Location | 7,180,871 – 7,180,963 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 67.11 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -14.45 |
| Energy contribution | -17.90 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.54 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7180871 92 + 22407834 ---CGGCUAACUGAUAAAAGUUAGCCACGUCUGGCUCCAUUAUUGGGAUGCCCCCAAAGGCCAAUC-GACAAACAAUUUCCGGAGCAUGGACUUC----------A ---.((((((((......))))))))..((((((((((....(((((.....))))).((..(((.-(.....).))).))))))).)))))...----------. ( -32.80) >DroSec_CAF1 122274 92 + 1 ---CGGCUAACUAAUAAAAGUUGGCCACGUCUGGCUCCAUUAUUGGGUUGCCCCCAAAGGCCAAUC-GACAAACAAUUUCCGGAGCAUGGACUUC----------A ---.((((((((......))))))))..((((((((((...((((.((((((......))).....-)))...))))....))))).)))))...----------. ( -31.70) >DroAna_CAF1 130715 98 + 1 AGCCAACCAACCGAUAAAAGUUGGCC-UAUCCAUCUCCAUUAUAGAGUACU-------UGCCAAUAGGGCAACGAAUUUCCGGGACAAGAAAUUAUAAAACCGAAA ..........(((..((..((((.((-(((.((.(((.......)))....-------))...))))).))))...))..)))....................... ( -16.30) >consensus ___CGGCUAACUGAUAAAAGUUGGCCACGUCUGGCUCCAUUAUUGGGUUGCCCCCAAAGGCCAAUC_GACAAACAAUUUCCGGAGCAUGGACUUC__________A ....((((((((......))))))))..((((((((((....(((((.....)))))...(......).............))))).))))).............. (-14.45 = -17.90 + 3.45)


| Location | 7,180,871 – 7,180,963 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.11 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -19.37 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.43 |
| SVM RNA-class probability | 0.999895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7180871 92 - 22407834 U----------GAAGUCCAUGCUCCGGAAAUUGUUUGUC-GAUUGGCCUUUGGGGGCAUCCCAAUAAUGGAGCCAGACGUGGCUAACUUUUAUCAGUUAGCCG--- .----------...(((...(((((((.(((((.....)-))))..)).(((((.....)))))....)))))..)))..((((((((......)))))))).--- ( -33.90) >DroSec_CAF1 122274 92 - 1 U----------GAAGUCCAUGCUCCGGAAAUUGUUUGUC-GAUUGGCCUUUGGGGGCAACCCAAUAAUGGAGCCAGACGUGGCCAACUUUUAUUAGUUAGCCG--- .----------...(((...(((((((.(((((.....)-))))..))....(((....)))......)))))..)))..(((.((((......)))).))).--- ( -32.20) >DroAna_CAF1 130715 98 - 1 UUUCGGUUUUAUAAUUUCUUGUCCCGGAAAUUCGUUGCCCUAUUGGCA-------AGUACUCUAUAAUGGAGAUGGAUA-GGCCAACUUUUAUCGGUUGGUUGGCU ((((((..(...........)..))))))((((((((((.....))))-------....((((.....)))))))))).-((((((((..........)))))))) ( -24.50) >consensus U__________GAAGUCCAUGCUCCGGAAAUUGUUUGUC_GAUUGGCCUUUGGGGGCAACCCAAUAAUGGAGCCAGACGUGGCCAACUUUUAUCAGUUAGCCG___ ..............(((...((((((..........(((.....))).....(((....))).....))))))..)))..((((((((......)))))))).... (-19.37 = -20.17 + 0.79)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:26 2006