| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
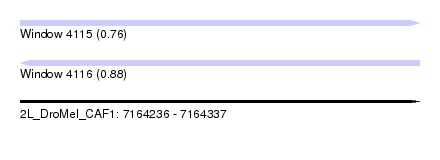
| Location | 7,164,236 – 7,164,337 |
| Length | 101 |
| Max. P | 0.882781 |

| Location | 7,164,236 – 7,164,337 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -15.92 |
| Energy contribution | -17.56 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7164236 101 + 22407834 AAUCAAAUUCAUUUGCCAACUGUUUUCACU--GCAAGCCAAAUGUGUC--UUUGGCUGACU-UUGGCA-ACCAGCGCUCA--------GAAGCC-----AUGGCAAAUGGAGCAGUCAGG .......(((((((((((..((.((((...--((.(((((((......--)))))))....-...((.-....))))...--------)))).)-----))))))))))))......... ( -29.00) >DroSec_CAF1 105616 98 + 1 AAUCAAAUUCAUUUGCCAACUGUUUUCACU--GCAAGCCAAAUGCGGC--UUUGGCUGACU----GCA-ACCAGCGCUCA--------GAAGCC-----AUGGCAAAUGGAGCAGUCAGG .......(((((((((((..((.((((...--(((((((......)))--))..((((...----...-..))))))...--------)))).)-----))))))))))))......... ( -30.80) >DroSim_CAF1 108721 98 + 1 AAUCAAAUUCAUUUGCCAACUGUUUUCACU--GCAAGCCAAAUGUGGC--UUUGGCUGACU----GCA-ACCAGCGCUCA--------GAAGCC-----AUGGCAAAUGGAGCAGUCAGG .......(((((((((((..((.((((...--((((((((....))))--))..((((...----...-..))))))...--------)))).)-----))))))))))))......... ( -31.50) >DroYak_CAF1 113674 107 + 1 AAUCAAAUUCAUUUGCAAACUGUUUUCACU--UCAAGCCAAAUCUGUC--U---GCUGUCUGCUGGUU-GCCAGCGCUCAGAAGCUCAGAAGCC-----UUGGCAAAUGGAGCAGUCAGG .......(((((((((....((....))..--.((((.(...((((.(--(---.(((..((((((..-.))))))..))).))..)))).).)-----))))))))))))......... ( -28.10) >DroAna_CAF1 110780 103 + 1 AAUCAAAUUCAUUUGCCAACAGUUUUCACUCGGCAAGCGAA-UCUGCCGGAUUGCCAGAUU----GCUUGGCAG----UA--------GAAGCUACCAAAUGGCAAAUGGAGCAACCAGG .......(((((((((((..(((((..(((.(.(((((.((-((((.(.....).))))))----))))).)))----).--------.)))))......)))))))))))......... ( -34.90) >consensus AAUCAAAUUCAUUUGCCAACUGUUUUCACU__GCAAGCCAAAUGUGGC__UUUGGCUGACU____GCA_ACCAGCGCUCA________GAAGCC_____AUGGCAAAUGGAGCAGUCAGG .......(((((((((((.................(((((((........)))))))........((......)).........................)))))))))))......... (-15.92 = -17.56 + 1.64)


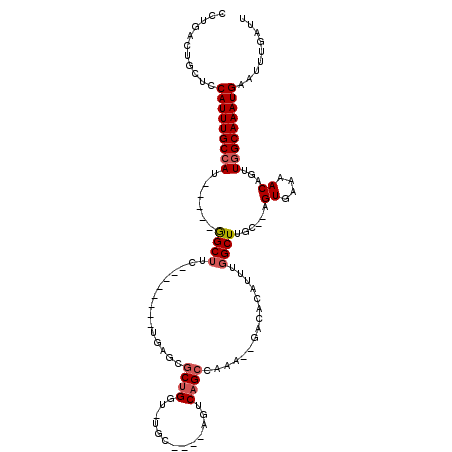
| Location | 7,164,236 – 7,164,337 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7164236 101 - 22407834 CCUGACUGCUCCAUUUGCCAU-----GGCUUC--------UGAGCGCUGGU-UGCCAA-AGUCAGCCAAA--GACACAUUUGGCUUGC--AGUGAAAACAGUUGGCAAAUGAAUUUGAUU ...........((((((((((-----(((..(--------(.......)).-.)))).-.((.(((((((--......))))))).))--.((....))...)))))))))......... ( -30.80) >DroSec_CAF1 105616 98 - 1 CCUGACUGCUCCAUUUGCCAU-----GGCUUC--------UGAGCGCUGGU-UGC----AGUCAGCCAAA--GCCGCAUUUGGCUUGC--AGUGAAAACAGUUGGCAAAUGAAUUUGAUU ...........(((((((((.-----.(((..--------..)))((((.(-..(----.((.(((((((--......))))))).))--.)..)...)))))))))))))......... ( -32.50) >DroSim_CAF1 108721 98 - 1 CCUGACUGCUCCAUUUGCCAU-----GGCUUC--------UGAGCGCUGGU-UGC----AGUCAGCCAAA--GCCACAUUUGGCUUGC--AGUGAAAACAGUUGGCAAAUGAAUUUGAUU ...........(((((((((.-----.(((..--------..)))((((.(-..(----.((.(((((((--......))))))).))--.)..)...)))))))))))))......... ( -32.50) >DroYak_CAF1 113674 107 - 1 CCUGACUGCUCCAUUUGCCAA-----GGCUUCUGAGCUUCUGAGCGCUGGC-AACCAGCAGACAGC---A--GACAGAUUUGGCUUGA--AGUGAAAACAGUUUGCAAAUGAAUUUGAUU ...........((((((((((-----(.(.((((..((.(((...(((((.-..)))))...))).---)--).))))...).)))).--.((....)).....)))))))......... ( -29.50) >DroAna_CAF1 110780 103 - 1 CCUGGUUGCUCCAUUUGCCAUUUGGUAGCUUC--------UA----CUGCCAAGC----AAUCUGGCAAUCCGGCAGA-UUCGCUUGCCGAGUGAAAACUGUUGGCAAAUGAAUUUGAUU ((.(((((((....((((...(((((((....--------..----)))))))))----))...))))))).))((((-((((.(((((((((....)))..)))))).))))))))... ( -39.90) >consensus CCUGACUGCUCCAUUUGCCAU_____GGCUUC________UGAGCGCUGGU_UGC____AGUCAGCCAAA__GACACAUUUGGCUUGC__AGUGAAAACAGUUGGCAAAUGAAUUUGAUU ...........(((((((((......((((...............((((.............))))...............))))......((....))...)))))))))......... (-16.13 = -16.77 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:11 2006