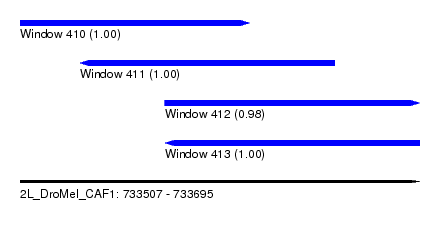
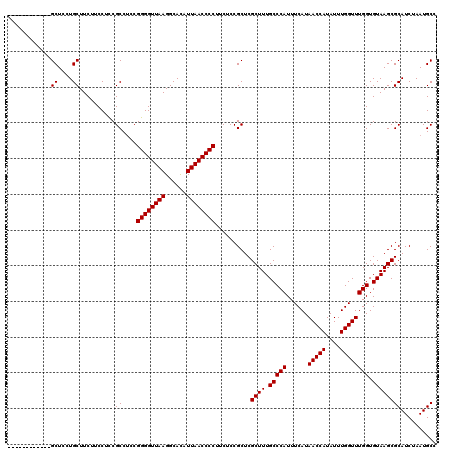
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 733,507 – 733,695 |
| Length | 188 |
| Max. P | 0.999795 |

| Location | 733,507 – 733,615 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -26.90 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 733507 108 + 22407834 ------------GCUCCUGCUUCUUCCUCCGCCUUCGGGGUUAAGGCACAUUAACCCCUUCUCCGCUCGCUUUGCCCAUUUCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCC ------------((....))..........((....((((((((......))))))))..........((((.(((((......(((((....)))))))).)))))))).......... ( -26.90) >DroSec_CAF1 5020 108 + 1 ------------GCUCCUGCUUCUUCCUCCGCCUCCGGGGUUAAGGCACAUUAACCCCUUCUCCGCUCGCUUUGCCCAUUUCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCC ------------((....))..........((....((((((((......))))))))..........((((.(((((......(((((....)))))))).)))))))).......... ( -26.90) >DroSim_CAF1 5019 108 + 1 ------------GCUCCUGCUUCUUCCUCCGCCUCCGGGGUUAAGGCACAUUAACCCCUUCUCCGCUCGCUUUGCCCAUUUCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCC ------------((....))..........((....((((((((......))))))))..........((((.(((((......(((((....)))))))).)))))))).......... ( -26.90) >DroEre_CAF1 5140 108 + 1 ------------GCUCGUGCUUCUUCCUCCGCCGCCGGGGUUAAGGCAUAUUAACCCCUUCUCCGCUCGCUUUGCCCAUUUCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCC ------------((..((((................((((((((......))))))))..........((((.(((((......(((((....)))))))).)))))))))).....)). ( -28.30) >DroYak_CAF1 5345 120 + 1 UCUCCUGCCUCUGCUCGUGCUUCUUCCUUCGCCUCCGGGGUUAAGGCAUAUUAACCCCUUCUCCGCUCGCUUUGCCCAUUUCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCC ............((..((((................((((((((......))))))))..........((((.(((((......(((((....)))))))).)))))))))).....)). ( -28.30) >consensus ____________GCUCCUGCUUCUUCCUCCGCCUCCGGGGUUAAGGCACAUUAACCCCUUCUCCGCUCGCUUUGCCCAUUUCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCC ............((....))..........((....((((((((......))))))))..........((((.(((((......(((((....)))))))).)))))))).......... (-26.90 = -26.90 + 0.00)

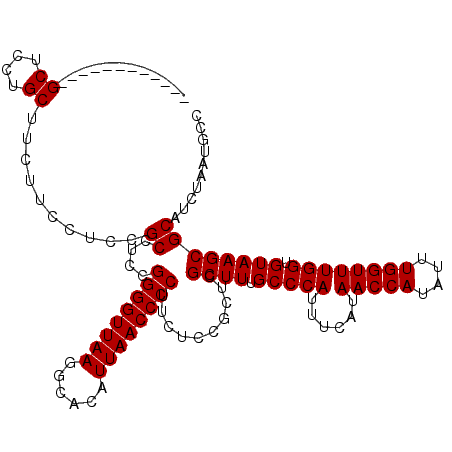
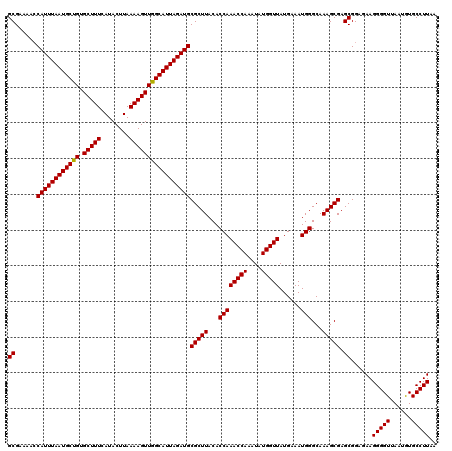
| Location | 733,535 – 733,655 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -33.88 |
| Energy contribution | -33.64 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 733535 120 - 22407834 GCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGAAAUGGGCAAAGCGAGCGGAGAAGGGGUUAAUGUGCCUUAA ((......((((((((((((.(((((........)))))))))))))))))(((((...((((((((....)))))......)))...))))).))......(((((......))))).. ( -32.50) >DroSec_CAF1 5048 120 - 1 GCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGAAAUGGGCAAAGCGAGCGGAGAAGGGGUUAAUGUGCCUUAA ((......((((((((((((.(((((........)))))))))))))))))(((((...((((((((....)))))......)))...))))).))......(((((......))))).. ( -32.50) >DroSim_CAF1 5047 120 - 1 GCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGAAAUGGGCAAAGCGAGCGGAGAAGGGGUUAAUGUGCCUUAA ((......((((((((((((.(((((........)))))))))))))))))(((((...((((((((....)))))......)))...))))).))......(((((......))))).. ( -32.50) >DroEre_CAF1 5168 120 - 1 GCGAAAAGCAUUUAAUGCCGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGAAAUGGGCAAAGCGAGCGGAGAAGGGGUUAAUAUGCCUUAA ((.....(((((((((((((.(((((........))))))))))))))))))((((...((((((((....)))))......)))...))))..))....((((.((....)).)))).. ( -38.50) >DroYak_CAF1 5385 120 - 1 GCGAAAACCAUUUAAUGCCGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGAAAUGGGCAAAGCGAGCGGAGAAGGGGUUAAUAUGCCUUAA ((......((((((((((((.(((((........)))))))))))))))))(((((...((((((((....)))))......)))...))))).))....((((.((....)).)))).. ( -35.60) >consensus GCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGAAAUGGGCAAAGCGAGCGGAGAAGGGGUUAAUGUGCCUUAA ((......((((((((((((.(((((........)))))))))))))))))(((((...((((((((....)))))......)))...))))).))......(((((......))))).. (-33.88 = -33.64 + -0.24)

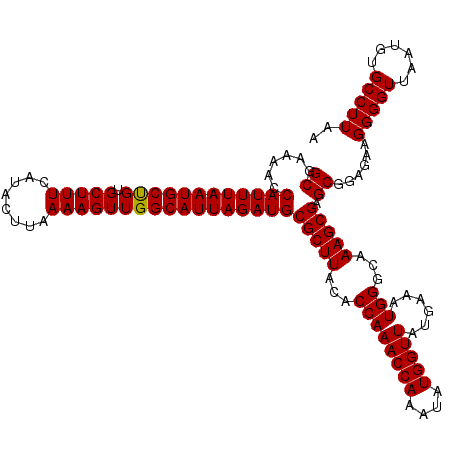
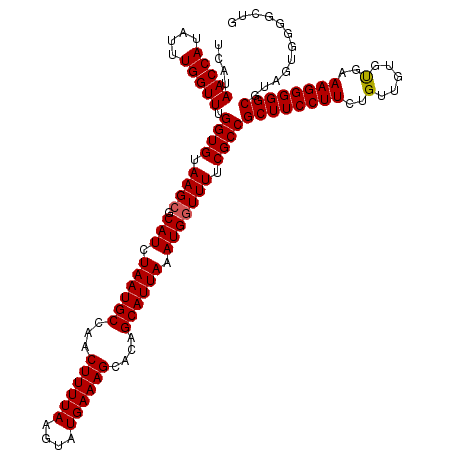
| Location | 733,575 – 733,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.12 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 733575 120 + 22407834 UCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCCAACUUUUAAGUAUGAAAGCACAGCAUUAAAUGGUUUUCGCCGCUUCCUUCUGUUGUGUGAAAGGGGGCGUAGUGGAGCUG (((((((((....))))).((((.((((.(((.((((((...((((((....))))))....)))))).))))))).))))(((((((((........).))))))))...))))..... ( -35.20) >DroSec_CAF1 5088 120 + 1 UCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCCAACUUUUAAGUAUGAAAGCACAGCAUUAAAUGGUUUUCGCCGCUUCCUUCUGUUGUGCGAAAGGGGGCGUAGGGGGCCCG ....(((((....))))).((((.((((.(((.((((((...((((((....))))))....)))))).))))))).))))((((((((.((.....)).))))))))...((....)). ( -38.20) >DroSim_CAF1 5087 120 + 1 UCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCCAACUUUUAAGUAUGAAAGCACAGCAUUAAAUGGUUUUCGCCGCUUCCUUCUGUUGUGCGAAAGGGGGCGUAGUGGGGCCG (((((((((....))))).((((.((((.(((.((((((...((((((....))))))....)))))).))))))).))))((((((((.((.....)).))))))))...))))..... ( -36.50) >DroEre_CAF1 5208 120 + 1 UCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCCAACUUUUAAGUAUGAAAGCACGGCAUUAAAUGCUUUUCGCCGCUUCCUUCUGUUGUGUGAAAGGGGGCGUAGUGGGGAUG .(((..((((.........((((.(((.((((.(((((((..((((((....))))))...))))))).)))).)))))))(((((((((........).))))))))...))))..))) ( -41.20) >DroYak_CAF1 5425 120 + 1 UCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCCAACUUUUAAGUAUGAAAGCACGGCAUUAAAUGGUUUUCGCCGCUUCCUUCUGUUGUGUGAAAGGGGGCGUAGUGGGGAUG .(((..((((.........((((.((((.(((.(((((((..((((((....))))))...))))))).))))))).))))(((((((((........).))))))))...))))..))) ( -41.20) >consensus UCAUAACCAUAUUUGGUUUGGUGUAAGCGCAUCUAAUGCCAACUUUUAAGUAUGAAAGCACAGCAUUAAAUGGUUUUCGCCGCUUCCUUCUGUUGUGUGAAAGGGGGCGUAGUGGGGCUG ....(((((....))))).((((.((((.(((.((((((...((((((....))))))....)))))).))))))).))))((((((((..(.....)..))))))))............ (-33.16 = -33.12 + -0.04)

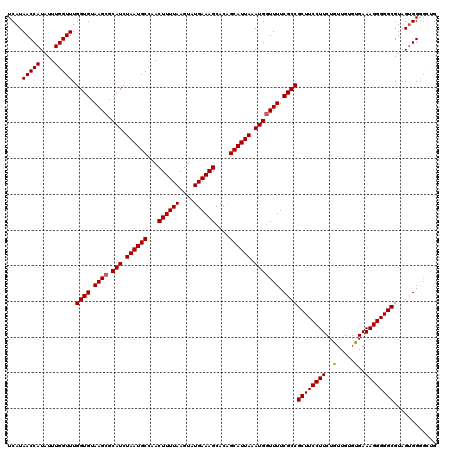
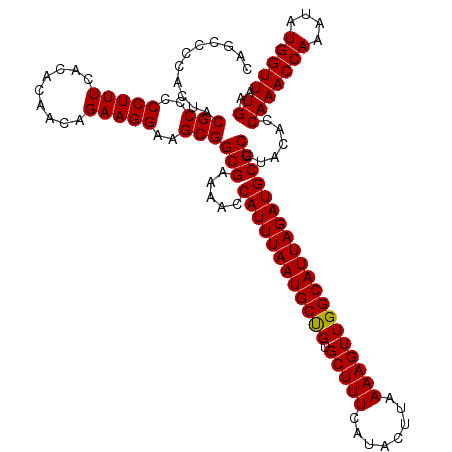
| Location | 733,575 – 733,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.36 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 733575 120 - 22407834 CAGCUCCACUACGCCCCCUUUCACACAACAGAAGGAAGCGGCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGA .(((.......(((((.(((((........)))))..).)))).....((((((((((((.(((((........))))))))))))))))).))).....(((((((....))))).)). ( -30.00) >DroSec_CAF1 5088 120 - 1 CGGGCCCCCUACGCCCCCUUUCGCACAACAGAAGGAAGCGGCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGA .((((.......)))).(((((........)))))(((((........((((((((((((.(((((........))))))))))))))))))))))....(((((((....))))).)). ( -32.00) >DroSim_CAF1 5087 120 - 1 CGGCCCCACUACGCCCCCUUUCGCACAACAGAAGGAAGCGGCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGA .((........(((..((((..(.....)..))))..)))(((.....((((((((((((.(((((........)))))))))))))))))))).....)).(((((....))))).... ( -30.50) >DroEre_CAF1 5208 120 - 1 CAUCCCCACUACGCCCCCUUUCACACAACAGAAGGAAGCGGCGAAAAGCAUUUAAUGCCGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGA (((..(((...(((((.(((((........)))))..).))))....(((((((((((((.(((((........))))))))))))))))))...................)))..))). ( -36.20) >DroYak_CAF1 5425 120 - 1 CAUCCCCACUACGCCCCCUUUCACACAACAGAAGGAAGCGGCGAAAACCAUUUAAUGCCGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGA (((..(((...(((..(((((.........)))))..)))(((.....((((((((((((.(((((........)))))))))))))))))))).................)))..))). ( -32.10) >consensus CAGCCCCACUACGCCCCCUUUCACACAACAGAAGGAAGCGGCGAAAACCAUUUAAUGCUGUGCUUUCAUACUUAAAAGUUGGCAUUAGAUGCGCUUACACCAAACCAAAUAUGGUUAUGA ...........(((..(((((.........)))))..)))(((.....((((((((((((.(((((........))))))))))))))))))))......(((((((....))))).)). (-29.60 = -29.36 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:23 2006