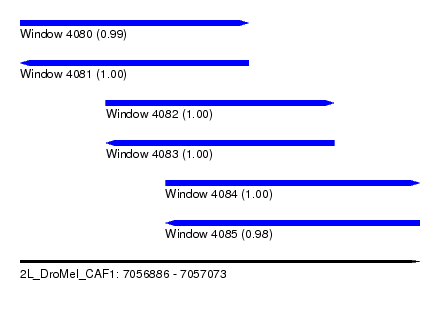
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,056,886 – 7,057,073 |
| Length | 187 |
| Max. P | 0.999991 |

| Location | 7,056,886 – 7,056,993 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7056886 107 + 22407834 UAAAGUACAAAGCGAAGAAAAGGAGAAAAUAAAUAAGCGGAAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAG ...........((((......((........((((((.......(.(((.....------------...))).)(((((((((.....)))).)))))..)))))).))....)))).. ( -21.00) >DroSec_CAF1 25499 107 + 1 UGAAGUACAAAGCGAAGAAAAGGAGAAAAUAAAUAAGCGGAAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAG ...........((((......((........((((((.......(.(((.....------------...))).)(((((((((.....)))).)))))..)))))).))....)))).. ( -21.00) >DroSim_CAF1 25085 107 + 1 UGAAGUACAAAGCGAAGAAAAGGAGAAAAUAAAUAAGCGGAAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAG ...........((((......((........((((((.......(.(((.....------------...))).)(((((((((.....)))).)))))..)))))).))....)))).. ( -21.00) >DroEre_CAF1 26348 119 + 1 UGAAGUACAAAGCGAAGAAAAGGAGAAAAUAAAUAAGCGGAAAAGGGCCGAGAAGGAGAAGGAGGAGAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAG ...........((((......................(((.......))).............((...((((..(((((((((.....)))).))))).))))....))....)))).. ( -20.60) >DroYak_CAF1 26401 107 + 1 UGAAGUACAAAGCGAAGAAAAGGAGAAAAUAAAUAAGCGGAAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAG ...........((((......((........((((((.......(.(((.....------------...))).)(((((((((.....)))).)))))..)))))).))....)))).. ( -21.00) >consensus UGAAGUACAAAGCGAAGAAAAGGAGAAAAUAAAUAAGCGGAAAAGGGCCGAGAA____________GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAG ...........((((......((........((((((.......(.(((....................))).)(((((((((.....)))).)))))..)))))).))....)))).. (-19.35 = -19.35 + -0.00)

| Location | 7,056,886 – 7,056,993 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.18 |
| SVM RNA-class probability | 0.999977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7056886 107 - 22407834 CUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUUCCGCUUAUUUAUUUUCUCCUUUUCUUCGCUUUGUACUUUA .(((.(((.((((((.((((((.....)))))).)))))).))).))).....------------....(((.......)))..................................... ( -18.30) >DroSec_CAF1 25499 107 - 1 CUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUUCCGCUUAUUUAUUUUCUCCUUUUCUUCGCUUUGUACUUCA .(((.(((.((((((.((((((.....)))))).)))))).))).))).....------------....(((.......)))..................................... ( -18.30) >DroSim_CAF1 25085 107 - 1 CUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUUCCGCUUAUUUAUUUUCUCCUUUUCUUCGCUUUGUACUUCA .(((.(((.((((((.((((((.....)))))).)))))).))).))).....------------....(((.......)))..................................... ( -18.30) >DroEre_CAF1 26348 119 - 1 CUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUCUCCUCCUUCUCCUUCUCGGCCCUUUUCCGCUUAUUUAUUUUCUCCUUUUCUUCGCUUUGUACUUCA .(((.(((.((((((.((((((.....)))))).)))))).))).))).....................(((.......)))..................................... ( -18.30) >DroYak_CAF1 26401 107 - 1 CUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUUCCGCUUAUUUAUUUUCUCCUUUUCUUCGCUUUGUACUUCA .(((.(((.((((((.((((((.....)))))).)))))).))).))).....------------....(((.......)))..................................... ( -18.30) >consensus CUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC____________UUCUCGGCCCUUUUCCGCUUAUUUAUUUUCUCCUUUUCUUCGCUUUGUACUUCA .(((.(((.((((((.((((((.....)))))).)))))).))).))).....................(((.......)))..................................... (-18.30 = -18.30 + 0.00)

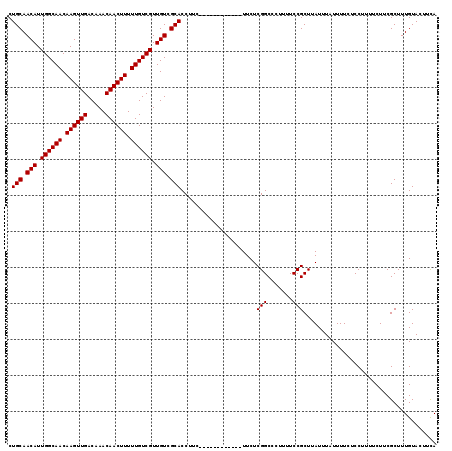
| Location | 7,056,926 – 7,057,033 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -38.25 |
| Energy contribution | -38.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.19 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7056926 107 + 22407834 AAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAAC ....(.(((.....------------...))).)(((((((((.....)))).)))))...(((((((........(((((((...)))))))(((((....))))).))))))).... ( -39.90) >DroSec_CAF1 25539 107 + 1 AAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAAC ....(.(((.....------------...))).)(((((((((.....)))).)))))...(((((((........(((((((...)))))))(((((....))))).))))))).... ( -39.90) >DroSim_CAF1 25125 107 + 1 AAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAAC ....(.(((.....------------...))).)(((((((((.....)))).)))))...(((((((........(((((((...)))))))(((((....))))).))))))).... ( -39.90) >DroEre_CAF1 26388 119 + 1 AAAAGGGCCGAGAAGGAGAAGGAGGAGAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAAC .......((.....)).......((...((((..(((((((((.....)))).))))).))))....))..((((((((((((...)))))).(((((....))))).))))))..... ( -38.40) >DroYak_CAF1 26441 107 + 1 AAAAGGGCCGAGAA------------GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAAC ....(.(((.....------------...))).)(((((((((.....)))).)))))...(((((((........(((((((...)))))))(((((....))))).))))))).... ( -39.90) >consensus AAAAGGGCCGAGAA____________GAAGGUGCGACAACGACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAAC ....(.(((....................))).)(((((((((.....)))).)))))...(((((((........(((((((...)))))))(((((....))))).))))))).... (-38.25 = -38.25 + 0.00)

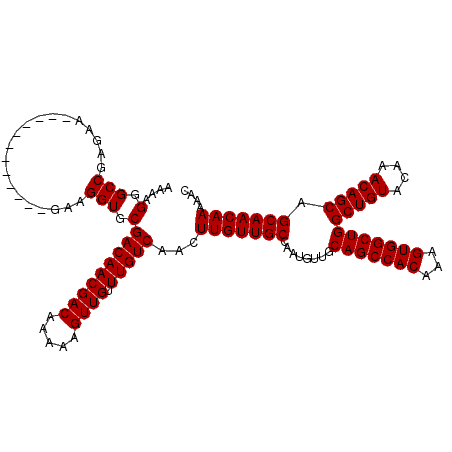
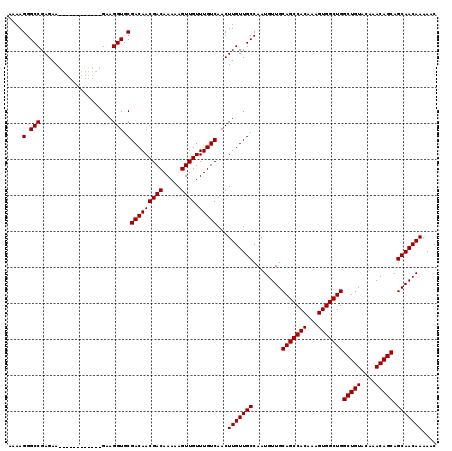
| Location | 7,056,926 – 7,057,033 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -32.50 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.65 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7056926 107 - 22407834 GUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUU .....((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).)))))...((((.......------------....))))...... ( -33.10) >DroSec_CAF1 25539 107 - 1 GUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUU .....((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).)))))...((((.......------------....))))...... ( -33.10) >DroSim_CAF1 25125 107 - 1 GUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUU .....((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).)))))...((((.......------------....))))...... ( -33.10) >DroEre_CAF1 26388 119 - 1 GUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUCUCCUCCUUCUCCUUCUCGGCCCUUUU .........((((((((....))))).((((((...))))))((.(((.((((((.((((((.....)))))).)))))).))).)).......................)))...... ( -32.50) >DroYak_CAF1 26441 107 - 1 GUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC------------UUCUCGGCCCUUUU .....((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).)))))...((((.......------------....))))...... ( -33.10) >consensus GUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUCGUUGUCGCACCUUC____________UUCUCGGCCCUUUU .........((((((((....))))).((((((...))))))((.(((.((((((.((((((.....)))))).)))))).))).)).......................)))...... (-32.50 = -32.50 + 0.00)

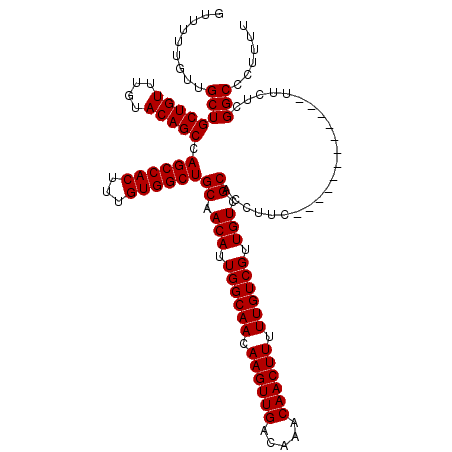
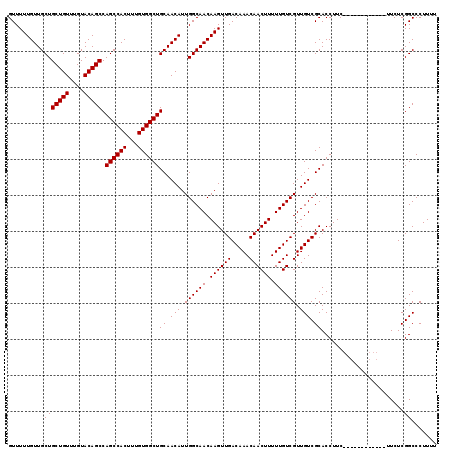
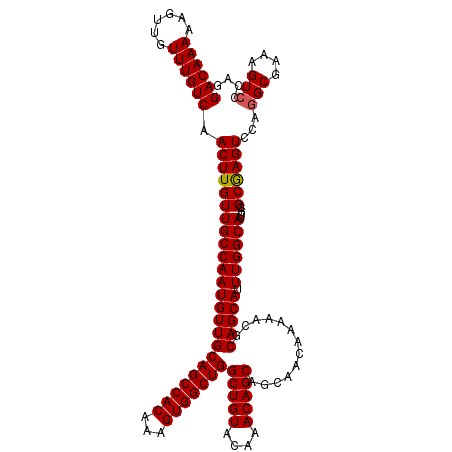
| Location | 7,056,954 – 7,057,073 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.66 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -43.90 |
| Energy contribution | -43.86 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7056954 119 + 22407834 GACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAACGCAGCAUUUGGCAUCGGGCAAGUCCAGGCGAAAGUACCAG ((((((......)))))).((((((((((((((((((((((((...)))))).(((((....)))))............)))))).))))))....))))))....((....))..... ( -44.40) >DroSec_CAF1 25567 119 + 1 GACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAACGCAGCAUUUGGCAUCGGGCGAGUCCAGGCGAAAGUCCCAG ((((((......)))))).((((((((((((((((((((((((...)))))).(((((....)))))............)))))).))))))....))))))...(((....))).... ( -46.00) >DroSim_CAF1 25153 119 + 1 GACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAACGCAGCAUUUGGCAUCGGGCGAGUCCAGGCGAAAGUCCCAG ((((((......)))))).((((((((((((((((((((((((...)))))).(((((....)))))............)))))).))))))....))))))...(((....))).... ( -46.00) >DroEre_CAF1 26428 119 + 1 GACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAACCCAGCAUUUGGCAUCGGGCGAGUCCAGGCGAAAGUCCCAG ((((((......)))))).((((((((((((((((((((((((...)))))))(((((....))))).............))))).))))))....))))))...(((....))).... ( -44.40) >DroYak_CAF1 26469 119 + 1 GACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAACCCAGCAUUUGGCAUCGGGCAAGUCCAGGCGAAAGUCCCAG ((((((......)))))).((((((((((((((((((((((((...)))))))(((((....))))).............))))).))))))....))))))...(((....))).... ( -44.70) >consensus GACAAAAAGUUGUUUGUCAACUUGUUGCCAAUGUUGCAGCCACAAAGUGGCUGGCUGUACAAACAGCAGCAACAAAAACGCAGCAUUUGGCAUCGGGCGAGUCCAGGCGAAAGUCCCAG ((((((......)))))).((((((((((((((((((((((((...)))))))(((((....))))).............))))).))))))....))))))...(((....))).... (-43.90 = -43.86 + -0.04)

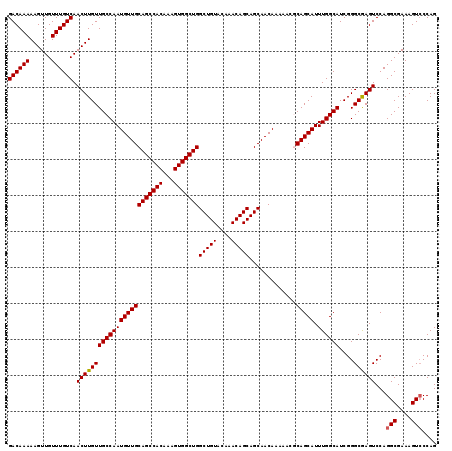
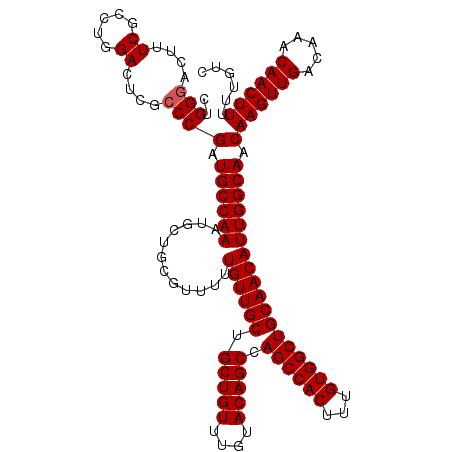
| Location | 7,056,954 – 7,057,073 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.66 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -34.74 |
| Energy contribution | -34.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7056954 119 - 22407834 CUGGUACUUUCGCCUGGACUUGCCCGAUGCCAAAUGCUGCGUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUC ..((((..(((....)))..)))).(((((........)))))..((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).))))) ( -36.60) >DroSec_CAF1 25567 119 - 1 CUGGGACUUUCGCCUGGACUCGCCCGAUGCCAAAUGCUGCGUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUC ..(((....((.....))....)))(((((........)))))..((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).))))) ( -37.70) >DroSim_CAF1 25153 119 - 1 CUGGGACUUUCGCCUGGACUCGCCCGAUGCCAAAUGCUGCGUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUC ..(((....((.....))....)))(((((........)))))..((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).))))) ( -37.70) >DroEre_CAF1 26428 119 - 1 CUGGGACUUUCGCCUGGACUCGCCCGAUGCCAAAUGCUGGGUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUC (..((.......))..)....(((((..((.....)))))))...((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).))))) ( -40.00) >DroYak_CAF1 26469 119 - 1 CUGGGACUUUCGCCUGGACUUGCCCGAUGCCAAAUGCUGGGUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUC (..((.......))..)....(((((..((.....)))))))...((((((.(((((....))))).((((((...))))))))))))..(((((.((((((.....)))))).))))) ( -39.90) >consensus CUGGGACUUUCGCCUGGACUCGCCCGAUGCCAAAUGCUGCGUUUUUGUUGCUGCUGUUUGUACAGCCAGCCACUUUGUGGCUGCAACAUUGGCAACAAGUUGACAAACAACUUUUUGUC ..(((....((.....))....)))(.((((((............((((((.(((((....))))).((((((...)))))))))))))))))).)((((((.....))))))...... (-34.74 = -34.94 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:41 2006