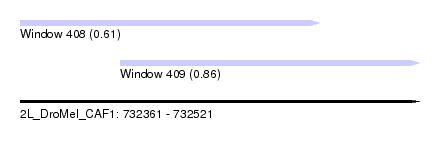
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 732,361 – 732,521 |
| Length | 160 |
| Max. P | 0.859824 |

| Location | 732,361 – 732,481 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -38.96 |
| Energy contribution | -39.68 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 732361 120 + 22407834 UUUUCCUGUGUGUCCGGAUGGAAAGGGGAUGGCGUUGAGAAUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGUGGCAGUGCAUCAUCCACUUGGCCAUUU ...((((.(...(((....))).).))))((((...(((....((((((((..(((.....(((((((((..((.....))..)))))).))))))...)).)))))).))).))))... ( -39.30) >DroSec_CAF1 3865 120 + 1 UUUUCCUCUGUGUCCGGAUGGAAAGGGGAUGGCAUUGAGAAUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUU ...((((((...(((....))).))))))((((...(((....((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).))).))))... ( -46.20) >DroSim_CAF1 3871 120 + 1 UUUUCCUCUGUGUCCGGAUGGAAAGGGGAUGGCGUUGAGAAUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUU ...((((((...(((....))).))))))((((...(((....((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).))).))))... ( -45.10) >DroEre_CAF1 3950 120 + 1 UUUCCCGCCGUGUCCGGAUGGAAAGGGGAUGGCGUUGAGAAUGGGAUGGGUUGGCCUGAGGACGGACCGGGGAUGGACUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUU ......(((((.(((.........))).)))))((..((....((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).))..))..... ( -44.40) >DroYak_CAF1 4159 120 + 1 UUUUCCUCCGUGUCCGGAUGGAAAGGGGAUGGCGUUGAGAAUGGGAUGGGUUGGCCCGAGGACGGACCGGGGAUGGACUGCAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUU ((((((((((....)))).)))))).(((((((...(((....((((.......((((.(......)))))((((.(((((..(.......)..))))).)))))))).))).))))))) ( -43.50) >consensus UUUUCCUCUGUGUCCGGAUGGAAAGGGGAUGGCGUUGAGAAUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUU ((((((((((....)))).)))))).(((((((...(((....((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).))).))))))) (-38.96 = -39.68 + 0.72)

| Location | 732,401 – 732,521 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -45.56 |
| Consensus MFE | -40.28 |
| Energy contribution | -41.80 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 732401 120 + 22407834 AUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGUGGCAGUGCAUCAUCCACUUGGCCAUUUGCCUGAUUGCGGUAUUUUGCUUCGGUCAGCUGGGUGGUGG ...((((((((..(((.....(((((((((..((.....))..)))))).))))))...)).)))))).....(((((..(.(((((((.(((.....))).))))))).)..))))).. ( -44.90) >DroSec_CAF1 3905 120 + 1 AUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUUGCCUGAUUGCGGUAUUUUGCCUCGGUCAGCUGGGUGGUGG ...((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).....(((((..(.(((((((.(((.....))).))))))).)..))))).. ( -49.80) >DroSim_CAF1 3911 120 + 1 AUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUUGCCUGAUUGCGGUAUUUUGCCUCGGUCAGCUGGGUGGUGG ...((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).....(((((..(.(((((((.(((.....))).))))))).)..))))).. ( -49.80) >DroEre_CAF1 3990 108 + 1 AUGGGAUGGGUUGGCCUGAGGACGGACCGGGGAUGGACUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUUGCCUGAUUGCGGUAUUUUGCUUCGGUCA------------ ...((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).))))))....(((.....))).(((((.(((.....))).))))).------------ ( -36.10) >DroYak_CAF1 4199 120 + 1 AUGGGAUGGGUUGGCCCGAGGACGGACCGGGGAUGGACUGCAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUUGCCUGAUUGCGGUAUUUUGCCUCGGUCAGCUGGGUGGUGG ..(((.(((((...((((.(......)))))((((.(((((..(.......)..))))).)))))))))))).(((((..(.(((((((.(((.....))).))))))).)..))))).. ( -47.20) >consensus AUGGGAUGGGCUGGCCUGAGGACGGACCGGGGAUGGAUUGUAACUGGUCAUGAGGCAGUGCAUCAUCCACUUGGCCAUUUGCCUGAUUGCGGUAUUUUGCCUCGGUCAGCUGGGUGGUGG ...((((((((..((((.(.....((((((..((.....))..)))))).).))))...)).)))))).....(((((..(.(((((((.(((.....))).))))))).)..))))).. (-40.28 = -41.80 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:19 2006