| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,040,905 – 7,041,002 |
| Length | 97 |
| Max. P | 0.787816 |

| Location | 7,040,905 – 7,041,002 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -19.23 |
| Energy contribution | -18.45 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
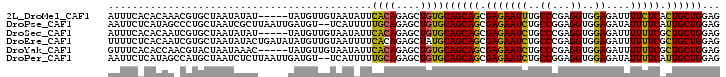
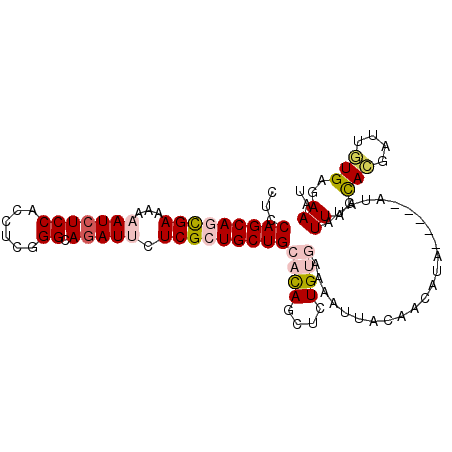
>2L_DroMel_CAF1 7040905 97 + 22407834 AUUUCACACAAACGUGCUAAUAUAU-----UAUGUUGUAAUAUUCACAGAGCUGUGCAGCAGCGAGAAUUUGCCCGAGGUGGAGAUUUUCUCACUGCUGGAG ......((((.....(((...((((-----(((...)))))))......)))))))((((((.(((((((..((...))..))....))))).))))))... ( -25.40) >DroPse_CAF1 12270 100 + 1 AAUUCUCAUAGCCCUGCUAAUCGCUUAAUUGAUGU--UCAUUUUUGCAGAGCUGUGCAGCAGCGAGAAUCUGCCGGAGGUGGAGAUAUUUUCAUUGCUGGAG ......((((((.((((..((((......))))..--........)))).))))))((((((.(((((((..((...))..))....))))).))))))... ( -28.60) >DroSec_CAF1 9527 97 + 1 AUUUCACACAAUCGUGCUAAUAUAU-----UAUGUUGUAAUAUUCACAGAGCUGUGCAGCAGCGAGAAUCUGCCCGAGGUGGAGAUUUUUUCGCUGCUGGAG ......((((.(((((.....((((-----(((...))))))).))).))..))))((((((((((((((..((...))..))....))))))))))))... ( -31.50) >DroEre_CAF1 9953 102 + 1 UUUUCUCACAAUCGUGCUAAUAUACUGAUAUAUGUUGUAAUUUUCACAGAGCUAUGCAGCAGCGAGAAUCUGCCCGAGGUGGAGAUUUUUUCGCUGCUGGAG ....(((......(((..(((.(((...........))))))..))).........((((((((((((((..((...))..))....))))))))))))))) ( -29.00) >DroYak_CAF1 9781 97 + 1 GUUUCACACCAACGUACUAAUAAAC-----UAUGUUGUAAUAUUCACAGAGCUGUGCAGCAGCGAGAAUCUGCCCGAGGUGGAGAUUUUUUCGCUGCUGGAG .........(((((((.........-----))))))).......((((....))))((((((((((((((..((...))..))....))))))))))))... ( -30.60) >DroPer_CAF1 11884 100 + 1 AAUUCUCAUAGCCAUGCUAAUCUCUUAAUUGAUGU--UCAUUUUUGCAGAGCUGUGCAGCAGCGAGAAUCUGCCGGAGGUGGAGAUAUUUUCAUUGCUGGAG ......((((((..(((..(((........)))..--........)))..))))))((((((.(((((((..((...))..))....))))).))))))... ( -26.30) >consensus AUUUCACACAAACGUGCUAAUAUAU_____UAUGUUGUAAUAUUCACAGAGCUGUGCAGCAGCGAGAAUCUGCCCGAGGUGGAGAUUUUUUCACUGCUGGAG ............................................((((....))))((((((.(((((((..((...))..))....))))).))))))... (-19.23 = -18.45 + -0.78)
| Location | 7,040,905 – 7,041,002 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -26.75 |
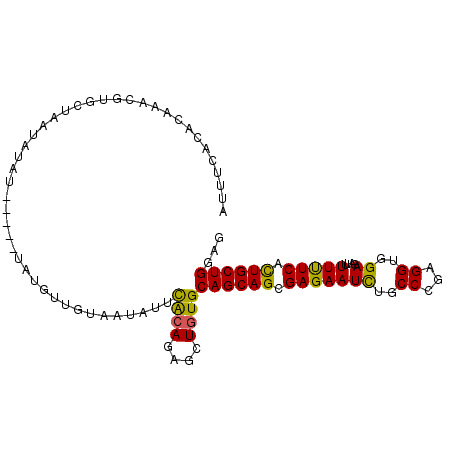
| Consensus MFE | -17.78 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7040905 97 - 22407834 CUCCAGCAGUGAGAAAAUCUCCACCUCGGGCAAAUUCUCGCUGCUGCACAGCUCUGUGAAUAUUACAACAUA-----AUAUAUUAGCACGUUUGUGUGAAAU ...((((((((((((....(((.....)))....))))))))))))((((....)))).((((((.....))-----))))....((((....))))..... ( -28.00) >DroPse_CAF1 12270 100 - 1 CUCCAGCAAUGAAAAUAUCUCCACCUCCGGCAGAUUCUCGCUGCUGCACAGCUCUGCAAAAAUGA--ACAUCAAUUAAGCGAUUAGCAGGGCUAUGAGAAUU ((((((((.(((....((((((......)).))))..))).)))))...((((((((.....((.--....))..(((....)))))))))))..))).... ( -26.10) >DroSec_CAF1 9527 97 - 1 CUCCAGCAGCGAAAAAAUCUCCACCUCGGGCAGAUUCUCGCUGCUGCACAGCUCUGUGAAUAUUACAACAUA-----AUAUAUUAGCACGAUUGUGUGAAAU ...(((((((((...(((((((......)).))))).)))))))))(((((.(((((.(((((.........-----..))))).))).)))))))...... ( -28.60) >DroEre_CAF1 9953 102 - 1 CUCCAGCAGCGAAAAAAUCUCCACCUCGGGCAGAUUCUCGCUGCUGCAUAGCUCUGUGAAAAUUACAACAUAUAUCAGUAUAUUAGCACGAUUGUGAGAAAA ...(((((((((...(((((((......)).))))).)))))))))(((((.((.(((..((((((...........))).)))..))))))))))...... ( -27.10) >DroYak_CAF1 9781 97 - 1 CUCCAGCAGCGAAAAAAUCUCCACCUCGGGCAGAUUCUCGCUGCUGCACAGCUCUGUGAAUAUUACAACAUA-----GUUUAUUAGUACGUUGGUGUGAAAC ...(((((((((...(((((((......)).))))).)))))))))((((.(..((((......((......-----)).......))))..).)))).... ( -28.82) >DroPer_CAF1 11884 100 - 1 CUCCAGCAAUGAAAAUAUCUCCACCUCCGGCAGAUUCUCGCUGCUGCACAGCUCUGCAAAAAUGA--ACAUCAAUUAAGAGAUUAGCAUGGCUAUGAGAAUU (((.(((.(((.....(((((........(((((.....((((.....))))))))).....((.--....)).....)))))...))).)))..))).... ( -21.90) >consensus CUCCAGCAGCGAAAAAAUCUCCACCUCGGGCAGAUUCUCGCUGCUGCACAGCUCUGUGAAAAUUACAACAUA_____AUAUAUUAGCACGAUUGUGAGAAAU ...(((((((((...(((((((......)).))))).)))))))))((((....))))........................((..(((....)))..)).. (-17.78 = -18.70 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:28 2006