
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,034,983 – 7,035,124 |
| Length | 141 |
| Max. P | 0.962689 |

| Location | 7,034,983 – 7,035,086 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
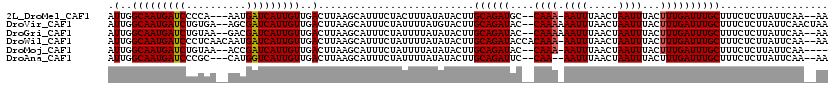
>2L_DroMel_CAF1 7034983 103 + 22407834 ---CUAU-A-UCC-AUAUUACUACCU----UUGCUAAAAAUUGGCAAUGAUCCCCA---AAUGAUCAUUGUUGACUUAAGCAUUUCUACUUUAUAUACUUGCAGAUGC--CAAA-AAUU ---....-.-...-((((........----.((((.....(..(((((((((....---...)))))))))..)....))))..........))))....((....))--....-.... ( -16.41) >DroGri_CAF1 3876 107 + 1 UUA-UAU-U-ACUACUACUGCUCUA-----UUGCUAUAAAUUGGCAAUGAUCUGUAA--GACGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC--CAAAAAAUU ...-...-.-.......((((....-----.((((.(((.(..(((((((((.....--...)))))))))..).)))))))....(((.....)))...))))....--......... ( -22.30) >DroWil_CAF1 6096 109 + 1 ---CUAUUA-UCU-CUAUUAUCUACU----UUGCUCUAAAUUGGCAAUGAUCCCUCAACAAUGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUACCACAAA-AAUU ---......-...-....(((((...----.((((.(((.(..(((((((((..........)))))))))..).)))))))....(((.....))).....))))).......-.... ( -18.00) >DroMoj_CAF1 3826 106 + 1 UUACUAU-U-ACU-CUAUUGCUAUA-----UUGCUGUAAAUUGGCAAUGAUCUGUAA--ACCGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC--CAAA-AAUU .......-.-...-...((((((((-----(((((.(((.(..(((((((((.(...--.).)))))))))..).)))))))..........)))))...))))....--....-.... ( -20.50) >DroAna_CAF1 3481 100 + 1 -----AU-ACUCU-CUAUUACCAACU-----UGCUAUAAAUUGGCAAUGAUCCCGC---CAUGGUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUUC--CAA--AAUU -----..-.....-............-----((((.(((.(..(((((((((....---...)))))))))..).)))))))..........................--...--.... ( -17.30) >DroPer_CAF1 4493 107 + 1 ---CUAU-A-UCU-CUCUGUCUCUCUAUUAUGGCUAUAAAUUGGCAAUGAUCCCCC----AUGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC-ACAAA-AAUU ---....-.-...-.(((((....(......)(((.(((.(..(((((((((....----..)))))))))..).))))))...................)))))...-.....-.... ( -20.20) >consensus ___CUAU_A_UCU_CUAUUACUAUCU____UUGCUAUAAAUUGGCAAUGAUCCCUA___AAUGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC__CAAA_AAUU ...............................((((.(((.(..(((((((((..........)))))))))..).)))))))..................................... (-16.32 = -16.52 + 0.19)


| Location | 7,035,012 – 7,035,124 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -18.69 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7035012 112 + 22407834 AUUGGCAAUGAUCCCCA---AAUGAUCAUUGUUGACUUAAGCAUUUCUACUUUAUAUACUUGCAGAUGC--CAAA-AAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAA--AA .(..(((((((((....---...)))))))))..)...(((((..((..............((....))--...(-((((.....))))).....))..)))))............--.. ( -19.30) >DroVir_CAF1 5129 116 + 1 AUUGGCAAUGAUCUGUGA--AGCGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUGUACUUGCAGAUAC--CAAAAAAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAACUAA .(..(((((((((.....--...)))))))))..).....((((.........))))....((((((..--....(((((.....)))))......)))))).................. ( -19.90) >DroGri_CAF1 3907 114 + 1 AUUGGCAAUGAUCUGUAA--GACGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC--CAAAAAAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAA--AA .(..(((((((((.....--...)))))))))..)..........................((((((..--....(((((.....)))))......))))))..............--.. ( -19.30) >DroWil_CAF1 6126 117 + 1 AUUGGCAAUGAUCCCUCAACAAUGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUACCACAAA-AAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAA--AA .(..(((((((((..........)))))))))..)..........................((((((.......(-((((.....)))))......))))))..............--.. ( -17.22) >DroMoj_CAF1 3857 111 + 1 AUUGGCAAUGAUCUGUAA--ACCGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC--CAAA-AAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAA---- .(..(((((((((.(...--.).)))))))))..)..........................((((((..--...(-((((.....)))))......))))))..............---- ( -19.00) >DroAna_CAF1 3508 111 + 1 AUUGGCAAUGAUCCCGC---CAUGGUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUUC--CAA--AAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAA--AA .(..(((((((((....---...)))))))))..)..........................(((((((.--..(--((((.....))))).....)))))))..............--.. ( -17.40) >consensus AUUGGCAAUGAUCCCUAA__AACGAUCAUUGUUGACUUAAGCAUUUCUAUUUUAUAUACUUGCAGAUAC__CAAA_AAUUUAACUAAUUUACUUUGAUUUGCUUUCUCUUAUUCAA__AA .(..(((((((((..........)))))))))..)..........................((((((....((((.((((.....))))...)))))))))).................. (-16.41 = -16.43 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:26 2006