
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,026,842 – 7,026,962 |
| Length | 120 |
| Max. P | 0.824282 |

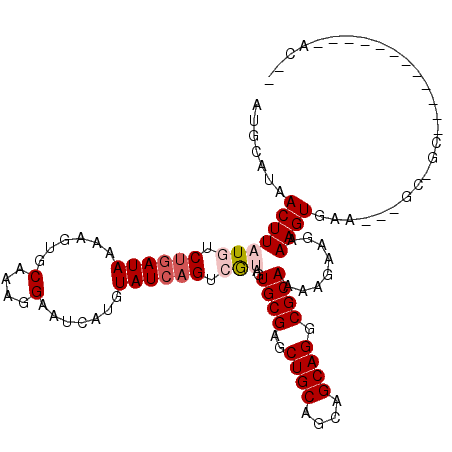
| Location | 7,026,842 – 7,026,933 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7026842 91 - 22407834 ACGCAUAACUUAUGUCUGAUAAAAGUGCGAAGGAAUCAUGUAUCAGUAAGAGUGCGAGCUGCAGCAGCAGGCGCAAGAGAAGAAAGUGAG---------------------C-- .(((...((((.((.((((((...((((....)...))).)))))))).))))(((..((((....)))).)))...........)))..---------------------.-- ( -23.00) >DroPse_CAF1 14797 103 - 1 AUGUAUAACUUAUGUCUGAUAAAAGUGCAAAGGAAUCCUGUAUCAGUCGUAGUGCGAGCUGCAGCAGCAGGCGCAAAAGAAGAAAGUGAAUUCGC-GC---------AGCAGG- .(((......((((.((((((.........(((...))).)))))).))))(((((((((((....)))).(((...........)))..)))))-))---------.)))..- ( -25.80) >DroGri_CAF1 122694 110 - 1 AUGUAGAACUUAUGUCUGAUACAAGUGCGCUGGAAUUAUGUAUAUGUCGUAGUGCGAGCUGCAGUAGCAGGCGCAA-AGAAGAAAGUGAAGCUGCUGCAUCAAAACC---AACA .((((..((....))....)))).....(((.(.((((((.......)))))).).)))(((((((((...(((..-........)))..)))))))))........---.... ( -26.70) >DroYak_CAF1 13313 91 - 1 ACGCAUAACUUAUGUCUGAUAAAAGUGCAAAGGAAUCAUGUAUCAGUAAGAGUGCGAGCUGCAGCAGCAGGCGCAAGAGAAGAAAGUGAG---------------------C-- .(((...((((.((.((((((...(((.........))).)))))))).))))(((..((((....)))).)))...........)))..---------------------.-- ( -21.80) >DroAna_CAF1 14649 99 - 1 AUGCAUAACUUAUGUCUGAUAAAAGUGCGAAGGAAUCAUGUAUCAGUCGUAGUGCGAUCUGCAGCAGCAGGCGCAAAAGAAGAAAGUGAG---GC-GU---------GGAGU-- ((((...(((((((.((((((...((((....)...))).)))))).)))..((((.(((((....)))))))))........))))...---))-))---------.....-- ( -28.90) >DroPer_CAF1 15762 103 - 1 AUGUAUAACUUAUGUCUGAUAAAAGUGCAAAGGAAUCCUGUAUCAGUCGUAGUGCGAGCUGCAGCAGCAGGCGCAAAAGAAGAAAGUGAAUUCGC-GC---------AGCAGG- .(((......((((.((((((.........(((...))).)))))).))))(((((((((((....)))).(((...........)))..)))))-))---------.)))..- ( -25.80) >consensus AUGCAUAACUUAUGUCUGAUAAAAGUGCAAAGGAAUCAUGUAUCAGUCGUAGUGCGAGCUGCAGCAGCAGGCGCAAAAGAAGAAAGUGAA___GC_GC____________AC__ .......(((((((.((((((......(....).......)))))).)))..((((..((((....)))).))))........))))........................... (-16.12 = -16.90 + 0.78)


| Location | 7,026,856 – 7,026,962 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -14.98 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7026856 106 + 22407834 CUUGCGCCUGCUGCUGCAGCUCGCACUCUUACUGAUACAUGAUUCCUUCGCACUUUUAUCAGACAUAAGUUAUGCGUAUGUUGGCAAGCACACUUUUUUCACGAAA- ..((.((.((((...(((...((((..((((((((((..(((.....)))......))))))...))))...))))..))).)))).)).))..............- ( -21.40) >DroVir_CAF1 100088 94 + 1 UUUGCGCCUGCUACUGCAGCUCGCACUACAACUGAUACAUAAUUCCAGCGCACCUGUAUCAGACAUAAGUUCUACAUAUGUUU-------------UUGCACAACUC ..((((.((((....))))..))))......((((((((...............))))))))(((((.(.....).)))))..-------------........... ( -20.06) >DroPse_CAF1 14823 94 + 1 UUUGCGCCUGCUGCUGCAGCUCGCACUACGACUGAUACAGGAUUCCUUUGCACUUUUAUCAGACAUAAGUUAUACAUAUGA------------UUUGUUCACUCAG- ..((((.((((....))))..))))......((((((.(((...........))).))))))..(((((((((....))))------------)))))........- ( -19.50) >DroMoj_CAF1 130221 94 + 1 UUUGCGCCUGCUACUGCAGCUCGCACUACAACUGAUACAUAAUUCCAGAGCACUUGUAUCAGACAUAAGUUCUACAUAUGUUU-------------UUGCACAAUUU ..((((.((((....))))..))))......((((((((...............))))))))(((((.(.....).)))))..-------------........... ( -20.86) >DroAna_CAF1 14671 104 + 1 UUUGCGCCUGCUGCUGCAGAUCGCACUACGACUGAUACAUGAUUCCUUCGCACUUUUAUCAGACAUAAGUUAUGCAUAUGCUGGACACCACAAUU--UGCACGAAA- ..((((.((((....))))..))))...((.((((((..(((.....)))......))))))..........((((..((.(((...)))))...--))))))...- ( -20.20) >DroPer_CAF1 15788 94 + 1 UUUGCGCCUGCUGCUGCAGCUCGCACUACGACUGAUACAGGAUUCCUUUGCACUUUUAUCAGACAUAAGUUAUACAUAUGA------------UUUGUUCACUCAG- ..((((.((((....))))..))))......((((((.(((...........))).))))))..(((((((((....))))------------)))))........- ( -19.50) >consensus UUUGCGCCUGCUGCUGCAGCUCGCACUACGACUGAUACAUGAUUCCUUCGCACUUUUAUCAGACAUAAGUUAUACAUAUGUU___________UU_UUGCACAAAA_ ..((((.((((....))))..))))......((((((...................)))))).((((.(.....).))))........................... (-14.98 = -14.98 + 0.00)



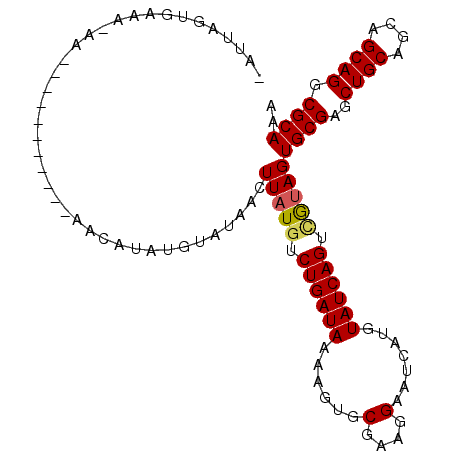
| Location | 7,026,856 – 7,026,962 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7026856 106 - 22407834 -UUUCGUGAAAAAAGUGUGCUUGCCAACAUACGCAUAACUUAUGUCUGAUAAAAGUGCGAAGGAAUCAUGUAUCAGUAAGAGUGCGAGCUGCAGCAGCAGGCGCAAG -..............(((((((((.......(((((..((((...((((((...((((....)...))).)))))))))).))))).((....)).))))))))).. ( -30.10) >DroVir_CAF1 100088 94 - 1 GAGUUGUGCAA-------------AAACAUAUGUAGAACUUAUGUCUGAUACAGGUGCGCUGGAAUUAUGUAUCAGUUGUAGUGCGAGCUGCAGUAGCAGGCGCAAA (((((.((((.-------------.......)))).)))))....((((((((....(....).....))))))))......((((..((((....)))).)))).. ( -27.10) >DroPse_CAF1 14823 94 - 1 -CUGAGUGAACAAA------------UCAUAUGUAUAACUUAUGUCUGAUAAAAGUGCAAAGGAAUCCUGUAUCAGUCGUAGUGCGAGCUGCAGCAGCAGGCGCAAA -....((((.....------------)))).........(((((.((((((.........(((...))).)))))).)))))((((..((((....)))).)))).. ( -21.80) >DroMoj_CAF1 130221 94 - 1 AAAUUGUGCAA-------------AAACAUAUGUAGAACUUAUGUCUGAUACAAGUGCUCUGGAAUUAUGUAUCAGUUGUAGUGCGAGCUGCAGUAGCAGGCGCAAA ......(((((-------------..(((((.(.....).)))))(((((((((((........))).))))))))))))).((((..((((....)))).)))).. ( -24.70) >DroAna_CAF1 14671 104 - 1 -UUUCGUGCA--AAUUGUGGUGUCCAGCAUAUGCAUAACUUAUGUCUGAUAAAAGUGCGAAGGAAUCAUGUAUCAGUCGUAGUGCGAUCUGCAGCAGCAGGCGCAAA -....(((((--...(((((...))).))..)))))...(((((.((((((...((((....)...))).)))))).)))))((((.(((((....))))))))).. ( -30.30) >DroPer_CAF1 15788 94 - 1 -CUGAGUGAACAAA------------UCAUAUGUAUAACUUAUGUCUGAUAAAAGUGCAAAGGAAUCCUGUAUCAGUCGUAGUGCGAGCUGCAGCAGCAGGCGCAAA -....((((.....------------)))).........(((((.((((((.........(((...))).)))))).)))))((((..((((....)))).)))).. ( -21.80) >consensus _AUUAGUGAAA_AA___________AACAUAUGUAUAACUUAUGUCUGAUAAAAGUGCGAAGGAAUCAUGUAUCAGUCGUAGUGCGAGCUGCAGCAGCAGGCGCAAA .......................................(((((.((((((......(....).......)))))).)))))((((..((((....)))).)))).. (-17.64 = -17.67 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:22 2006