
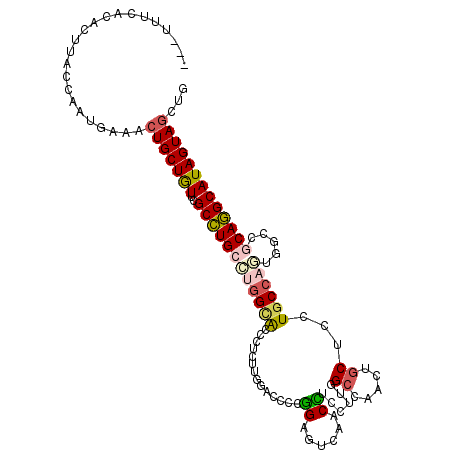
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,016,424 – 7,016,534 |
| Length | 110 |
| Max. P | 0.976255 |

| Location | 7,016,424 – 7,016,534 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.62 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7016424 110 + 22407834 ---AGCCACACUUACCAAUGAAACUGCUGUUCGCCUGCCUGGCACCCUCUUGGACCCCGGAGUGAACCUCCUUGGCCAACUGCUCCUGCCAGUGCCCGCAGGCAUAGUAGCUU ---....................(((((((..((((((..(((((....((((.((..((((.....))))..))))))..((....))..))))).)))))))))))))... ( -38.90) >DroVir_CAF1 87887 112 + 1 -UAUUUUGGACUCACCCAUCAAGGUGCUGUUUGCCUGCGUUGCGCCCUCUUGCACUCCGGCAGCCACCUCGGCGGCCGAUAGCGCCUGCCAUUUGCCACAGGCAUAGUAGCUG -.....(((......)))...((((((((((.(((((((..(.((......)).)..).)))(((.....)))))).))))))))))((.(((((((...)))).))).)).. ( -37.10) >DroGri_CAF1 108439 113 + 1 CCACUUUUUACAUACCAAUAAAGCUGCUGUUUGCCUGCCGGGUGCCAUCGAGCACACCGGCUGCAACUUGUGCGGCCAGCUGCUGUUGCCACUUGCCGCAGGCAUAGUAGCUG .....................((((((((..((((((((((((((......)))).)))))..........(((((.((..((....))..)).)))))))))))))))))). ( -47.90) >DroSim_CAF1 2743 110 + 1 ---AACCACACUUACCAAUGAAACUGCUGUUCGCCUGCCUGGCGCCCUCUUGAACCCCGGAGUGAACCUCCUUGGCCAACUGCUCCUGCCAGAGGCCGCAGGCAUAGUAGCUU ---....................(((((((..((((((..((..(.((((.(....).)))).)..)).....((((..((((....).))).)))))))))))))))))... ( -33.40) >DroYak_CAF1 2754 110 + 1 ---UUACCAACUUACCAAUGAAACUGCUGUUCGCCUGCCUGGCACCCUCUUGGACCCCGGAGUGCACCUCCUUGGCCAACUGCUCCUGCCAGCGCCCGCAGGCAUAGUAGCUU ---....................(((((((..((((((((((((..(..((((.((..((((.....))))..))))))..)....)))))).....)))))))))))))... ( -37.40) >DroAna_CAF1 3841 113 + 1 CUAUUUUUUACUUACUAAUAAAGCUGCUAUUAGCUUGACGGGAACCCUCCUGGACUCCAGCAUCCACUUCCUUUGCCAAAUCCUCUUGCCACUGACCACAAGCAUAGUAGCUG .....................(((((((((..(((((.(((((....))))).......(((...........)))......................)))))))))))))). ( -25.20) >consensus ___UUUCACACUUACCAAUGAAACUGCUGUUCGCCUGCCUGGCACCCUCUUGGACCCCGGAGUCAACCUCCUUGGCCAACUGCUCCUGCCAGUGGCCGCAGGCAUAGUAGCUG .......................(((((((..((((((((((((..............((......))......((.....))...)))))).....)))))))))))))... (-18.20 = -18.57 + 0.37)


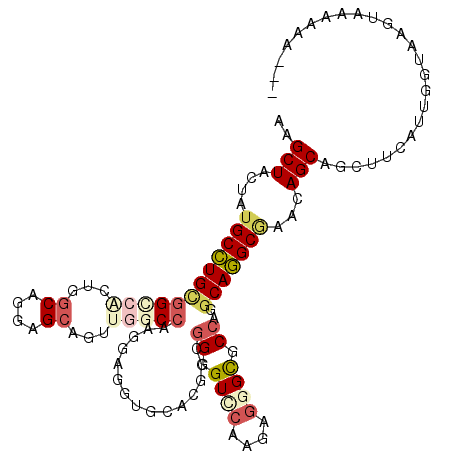
| Location | 7,016,424 – 7,016,534 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.62 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -21.32 |
| Energy contribution | -22.60 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7016424 110 - 22407834 AAGCUACUAUGCCUGCGGGCACUGGCAGGAGCAGUUGGCCAAGGAGGUUCACUCCGGGGUCCAAGAGGGUGCCAGGCAGGCGAACAGCAGUUUCAUUGGUAAGUGUGGCU--- .((((((...((((((.((((((.((....))..((((((..((((.....))))..)).))))...))))))..))))))((((....))))...........))))))--- ( -47.70) >DroVir_CAF1 87887 112 - 1 CAGCUACUAUGCCUGUGGCAAAUGGCAGGCGCUAUCGGCCGCCGAGGUGGCUGCCGGAGUGCAAGAGGGCGCAACGCAGGCAAACAGCACCUUGAUGGGUGAGUCCAAAAUA- ..(((....((((((((.....((((((.((((.((((...)))))))).))))))..((((......))))..)))))))).....(((((....))))))))........- ( -46.80) >DroGri_CAF1 108439 113 - 1 CAGCUACUAUGCCUGCGGCAAGUGGCAACAGCAGCUGGCCGCACAAGUUGCAGCCGGUGUGCUCGAUGGCACCCGGCAGGCAAACAGCAGCUUUAUUGGUAUGUAAAAAGUGG ....(((.(((((((((((.(((.((....)).))).)))))).(((((((.(((((.(((((....)))))))))).(.....).)))))))....))))))))........ ( -48.60) >DroSim_CAF1 2743 110 - 1 AAGCUACUAUGCCUGCGGCCUCUGGCAGGAGCAGUUGGCCAAGGAGGUUCACUCCGGGGUUCAAGAGGGCGCCAGGCAGGCGAACAGCAGUUUCAUUGGUAAGUGUGGUU--- .((((((..((((((((.(((((.((....))....((((..((((.....))))..))))..))))).)).)))))).((.....))................))))))--- ( -42.40) >DroYak_CAF1 2754 110 - 1 AAGCUACUAUGCCUGCGGGCGCUGGCAGGAGCAGUUGGCCAAGGAGGUGCACUCCGGGGUCCAAGAGGGUGCCAGGCAGGCGAACAGCAGUUUCAUUGGUAAGUUGGUAA--- .((((((((.((((((.((((((.((....))..((((((..((((.....))))..)).))))...))))))..))))))((((....))))...)))).)))).....--- ( -44.30) >DroAna_CAF1 3841 113 - 1 CAGCUACUAUGCUUGUGGUCAGUGGCAAGAGGAUUUGGCAAAGGAAGUGGAUGCUGGAGUCCAGGAGGGUUCCCGUCAAGCUAAUAGCAGCUUUAUUAGUAAGUAAAAAAUAG ..((((((..((.....)).))))))........(((((......(((....)))(((..((....))..))).)))))((((((((.....))))))))............. ( -25.50) >consensus AAGCUACUAUGCCUGCGGCCACUGGCAGGAGCAGUUGGCCAAGGAGGUGCACGCCGGGGUCCAAGAGGGCGCCAGGCAGGCGAACAGCAGCUUCAUUGGUAAGUAAAAAA___ ..(((....((((((((((((...((....))...)))))...............((.((((....)))).))..)))))))...)))......................... (-21.32 = -22.60 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:17 2006