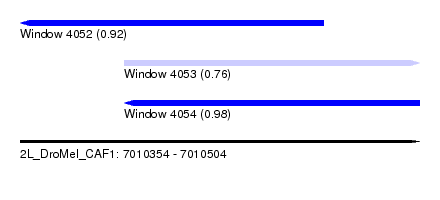
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,010,354 – 7,010,504 |
| Length | 150 |
| Max. P | 0.984263 |

| Location | 7,010,354 – 7,010,468 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -25.20 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7010354 114 - 22407834 GAAUUCGACGGCGGCGAAUAAC---GAUCGGUAUGCCGGUUGUUUUUUCGCCGACCGAUAACCAUAGAAAUGUGAAAAAAACCUAAGAAAUACGAAGCUUUUCGGGGCUUCAGUAUG ...(((..((((((((((.(((---((((((....)))))))))..))))))).))).....((((....))))............)))(((((((((((....))))))).)))). ( -44.70) >DroSec_CAF1 31294 112 - 1 GAAUUUAACGGCGGCGAAUAAC---GAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUACUAAUGUGAAA-AAACCUAAGAAAUCGGGAGCU-UUCGGGGCUUCAGUAUG ........((((((((((((((---((((((....))))))).)))))))))).))).....((((((...((....-..))...........((((((-.....)))))))))))) ( -42.40) >DroSim_CAF1 31072 113 - 1 GAAUUUAACGGCGGCGAAUAAC---GAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUACUAAUGUGAAAAAAACCUAAGAAAUCGGGAGCU-UUCGGGGCUUCAGUAUG ........((((((((((((((---((((((....))))))).)))))))))).))).....((((((.............((..(....)..))((((-.....))))..)))))) ( -42.40) >DroEre_CAF1 31311 111 - 1 GAAUUCAACGGCGGCGAAUAAC---GAUCGGUAUGCCGGCUGUUU-UUCACCGACCGAUAACCAUACUAAUGUGAAA-AAACCCAAGACAUCGGGAGCU-UUCGCGGCUUCAGUAUG ........((((((.(((.(((---(..(((....)))..)))).-))).))).))).....((((((..(((((((-...(((........)))...)-)))))).....)))))) ( -36.80) >DroYak_CAF1 31239 102 - 1 GAAUUCAACGGCGGCGAGUAACGACGAUCGGUAUGCCGGUUGUUU-UUCACCGA------------CUUGUGUGAAG-AAACCUAAGAAAUCGGAAGCU-UUCGGGGCUUCAGUAUG ...(((((((((((.(((....(((((((((....))))))))).-))).))).------------).))).))))(-((.(((..((((.(....).)-))).))).)))...... ( -30.80) >consensus GAAUUCAACGGCGGCGAAUAAC___GAUCGGUAUGCCGGUUGUUU_UUCGCCGACCGAUAACCAUACUAAUGUGAAA_AAACCUAAGAAAUCGGGAGCU_UUCGGGGCUUCAGUAUG ........((((((((((.......((((((....)))))).....))))))).)))....................................((((((......))))))...... (-25.20 = -26.00 + 0.80)


| Location | 7,010,393 – 7,010,504 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -25.42 |
| Energy contribution | -24.86 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7010393 111 + 22407834 UUUUCACAUUUCUAUGGUUAUCGGUCGGCGAAAAAACAACCGGCAUACCGAUC---GUUAUUCGCCGCCGUCGAAUUCGGCCGAAAUGAUGGCUCGGUAAUCUCAGCCGGCACU ....((((((((..........((.(((((((..(((...(((....)))...---))).)))))))))((((....)))).)))))).))((.((((.......))))))... ( -35.70) >DroSec_CAF1 31332 110 + 1 -UUUCACAUUAGUAUGGUUAUCGGUCGGCGAAUAAACAACCGGCAUACCGAUC---GUUAUUCGCCGCCGUUAAAUUCGGCCGAAAAGACGGCUCGGUAAUCUCAGCCGGCACU -.........(((.(((((..(((.((((((((((.....(((....)))...---.)))))))))))))......(((((((......)))).))).......)))))..))) ( -36.90) >DroSim_CAF1 31110 111 + 1 UUUUCACAUUAGUAUGGUUAUCGGUCGGCGAAUAAACAACCGGCAUACCGAUC---GUUAUUCGCCGCCGUUAAAUUCGGCCGAAAAGACGGCUCGGUAAUCUCAGCCGGCACU ..........(((.(((((..(((.((((((((((.....(((....)))...---.)))))))))))))......(((((((......)))).))).......)))))..))) ( -36.90) >DroEre_CAF1 31349 109 + 1 -UUUCACAUUAGUAUGGUUAUCGGUCGGUGAA-AAACAGCCGGCAUACCGAUC---GUUAUUCGCCGCCGUUGAAUUCGGCCGAAAAGACGGCUCGGUAAUCUCAACCGGCACU -.........(((.(((((....((((((...-.....)))))).((((((((---(((.((((..((((.......))))))))..))))).)))))).....)))))..))) ( -35.60) >DroYak_CAF1 31277 100 + 1 -CUUCACACAAG------------UCGGUGAA-AAACAACCGGCAUACCGAUCGUCGUUACUCGCCGCCGUUGAAUUCGGCCAAAAAGACGGCUCGGCAAUCUCAACCGGCACU -...........------------..(((((.-.(((.(((((....)))...)).)))..)))))((((((((..((((((........))).))).....)))).))))... ( -27.00) >consensus _UUUCACAUUAGUAUGGUUAUCGGUCGGCGAA_AAACAACCGGCAUACCGAUC___GUUAUUCGCCGCCGUUGAAUUCGGCCGAAAAGACGGCUCGGUAAUCUCAGCCGGCACU .......................((((((...........(((....))).............(((((((((...(((....)))..))))))..))).......))))))... (-25.42 = -24.86 + -0.56)


| Location | 7,010,393 – 7,010,504 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -30.60 |
| Energy contribution | -31.28 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7010393 111 - 22407834 AGUGCCGGCUGAGAUUACCGAGCCAUCAUUUCGGCCGAAUUCGACGGCGGCGAAUAAC---GAUCGGUAUGCCGGUUGUUUUUUCGCCGACCGAUAACCAUAGAAAUGUGAAAA .....((((((((((....((....)))))))))))).......((((((((((.(((---((((((....)))))))))..))))))).))).....((((....)))).... ( -46.60) >DroSec_CAF1 31332 110 - 1 AGUGCCGGCUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUUAACGGCGGCGAAUAAC---GAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUACUAAUGUGAAA- ((((.(((((((((..((......))..))))))))).......((((((((((((((---((((((....))))))).)))))))))).))).......)))).........- ( -47.10) >DroSim_CAF1 31110 111 - 1 AGUGCCGGCUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUUAACGGCGGCGAAUAAC---GAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUACUAAUGUGAAAA ((((.(((((((((..((......))..))))))))).......((((((((((((((---((((((....))))))).)))))))))).))).......)))).......... ( -47.10) >DroEre_CAF1 31349 109 - 1 AGUGCCGGUUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUCAACGGCGGCGAAUAAC---GAUCGGUAUGCCGGCUGUUU-UUCACCGACCGAUAACCAUACUAAUGUGAAA- ((((..(((((.((....((.((((......))))))...))..((((((.(((.(((---(..(((....)))..)))).-))).))).))).))))).)))).........- ( -37.50) >DroYak_CAF1 31277 100 - 1 AGUGCCGGUUGAGAUUGCCGAGCCGUCUUUUUGGCCGAAUUCAACGGCGGCGAGUAACGACGAUCGGUAUGCCGGUUGUUU-UUCACCGA------------CUUGUGUGAAG- ..(((((.(((((.((((((((.......))))).))).))))))))))((((((...(((((((((....))))))))).-.......)------------)))))......- ( -33.00) >consensus AGUGCCGGCUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUCAACGGCGGCGAAUAAC___GAUCGGUAUGCCGGUUGUUU_UUCGCCGACCGAUAACCAUACUAAUGUGAAA_ .....(((((((((..((......))..))))))))).......((((((((((.......((((((....)))))).....))))))).)))..................... (-30.60 = -31.28 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:11 2006