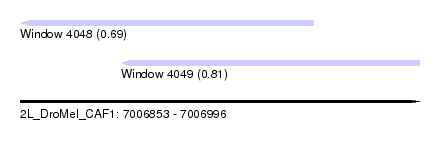
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,006,853 – 7,006,996 |
| Length | 143 |
| Max. P | 0.812218 |

| Location | 7,006,853 – 7,006,958 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
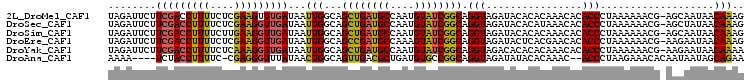
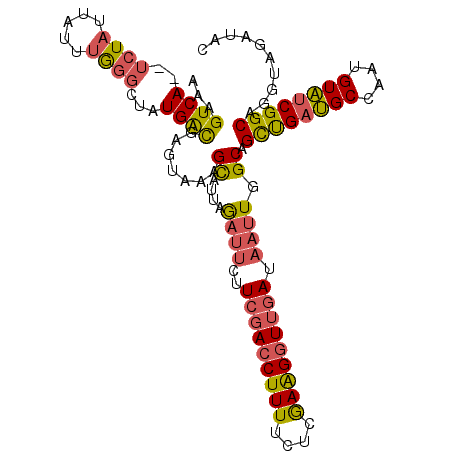
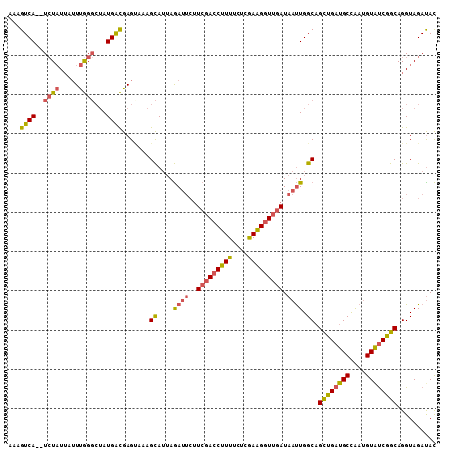
>2L_DroMel_CAF1 7006853 105 - 22407834 UAGAUUCUUCGACCUUUUCUCGAAGUUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUACACACAAACACACCCUAAAAAACG-AGCAAUAACAAAG (((....(((((.......)))))(((((...(((((((.....)))))))(((((........)))))...))))).....))).......-............. ( -22.50) >DroSec_CAF1 27858 105 - 1 UAGAUUCUUCGACCUUUUCUCGAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUACACAUAAACACACCCUAAAAAACG-AGCUAUAACAAAG (((.(((.(((((((((....)))))))))..(((((((.....)))))))(((((........)))))......................)-)))))........ ( -25.60) >DroSim_CAF1 27612 105 - 1 UAGAUUCUUCGACCUUUUCUUGAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUACACACAAACACACCCUAAAAAACG-AGCAAUAACAAAG (((.....(((((((((....)))))))))..(((((((.....)))))))(((((........))))).............))).......-............. ( -24.70) >DroEre_CAF1 27889 105 - 1 UAGAUUCUUCGACCUUUUCUCGAAGGUUGAUAAUUGGCAGCCGAUGCCAAAGUAUCGGCAGGUAGAUACUCACGAACACACCCUAAAAAACG-AAGAAUAACAAAG ...((((((((((((((....))))))(((..(((.((.((((((((....))))))))..)).)))..)))..................))-))))))....... ( -28.80) >DroYak_CAF1 27713 105 - 1 UAGAUUCUUCGACCUUUUCUCAAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGACACACACAAACACACCCUAAAAAACG-AAGAAUAACAAAA ...((((((((..((((....))))((((((((((((((.....))))))).)))))))...............................))-))))))....... ( -26.20) >DroAna_CAF1 25917 99 - 1 AAAA----UCUACCUUUUC-CGAGGGUUUAUAACUGGCAGUUGACGCUGAUGUGCCGGCAGGUAGAUAUACACAAAC--ACCCUAAGAAACACAAUAAUAGCAGAA ....----.......((((-..(((((......(((((((((......))).))))))...(((....)))......--)))))..))))................ ( -18.40) >consensus UAGAUUCUUCGACCUUUUCUCGAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUACACACAAACACACCCUAAAAAACG_AACAAUAACAAAG ........(((((((((....)))))))))...(((...((((((((....)))))))).(((................)))...................))).. (-19.86 = -19.42 + -0.44)
| Location | 7,006,889 – 7,006,996 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.65 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
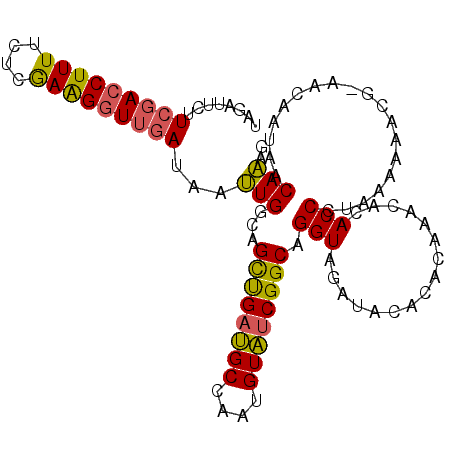
>2L_DroMel_CAF1 7006889 107 - 22407834 AAAGUCA--UCUAUUAUUUGGGCUAUGACGAGUUCAGCAUUAGAUUCUUCGACCUUUUCUCGAAGUUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUAC ......(--((((((...((((((......))))))((........((((((.......))))))..((((((((((((.....))))))).))))))).))))))).. ( -29.40) >DroSec_CAF1 27894 107 - 1 AAAGUCA--UAUAUUAUUUGGGCUAUGACGAGCUAAGCAUUAGAUUCUUCGACCUUUUCUCGAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUAC ...((((--((............))))))..(((..((....((((..(((((((((....))))))))).)))).)).((((((((....)))))))).)))...... ( -30.00) >DroSim_CAF1 27648 107 - 1 AAAGUCA--UCUAUUAUUUGGGCUAUGACGAGCUAAGCAUUAGAUUCUUCGACCUUUUCUUGAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUAC ......(--((((((.....((((......))))..((....((((..(((((((((....))))))))).)))).)).((((((((....)))))))).))))))).. ( -33.30) >DroEre_CAF1 27925 107 - 1 AAAGUCA--UCUAUUAUUUGGGCUAUGGCGAGAAAAGCAUUAGAUUCUUCGACCUUUUCUCGAAGGUUGAUAAUUGGCAGCCGAUGCCAAAGUAUCGGCAGGUAGAUAC ......(--((((((......((((..((.......))..........(((((((((....)))))))))....)))).((((((((....)))))))).))))))).. ( -33.60) >DroYak_CAF1 27749 107 - 1 AAAGUCA--UCUAUUAUUUGGGCUAUGACGAGUAAAGCAUUAGAUUCUUCGACCUUUUCUCAAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGACAC ...((((--(((.(((((((........))))))).((....((((..(((((((((....))))))))).)))).)).((((((((....)))))))))))).))).. ( -33.60) >DroAna_CAF1 25952 101 - 1 AAAGUCAGAUCUCUGAGUCAG---AUGAUGAGUAUUGUAAAAAA----UCUACCUUUUC-CGAGGGUUUAUAACUGGCAGUUGACGCUGAUGUGCCGGCAGGUAGAUAU ....((((....))))(((..---..)))..............(----(((((((..((-....)).......(((((((((......))).)))))).)))))))).. ( -21.50) >consensus AAAGUCA__UCUAUUAUUUGGGCUAUGACGAGUAAAGCAUUAGAUUCUUCGACCUUUUCUCGAAGGUUGAUAAUUGGCAGCUGAUGCCAAUGUAUCGGCAGGUAGAUAC ...((((..((((.....))))...)))).......((....((((..(((((((((....))))))))).)))).)).((((((((....)))))))).......... (-25.50 = -25.65 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:07 2006