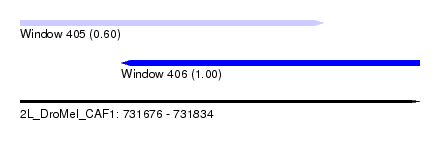
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 731,676 – 731,834 |
| Length | 158 |
| Max. P | 0.996655 |

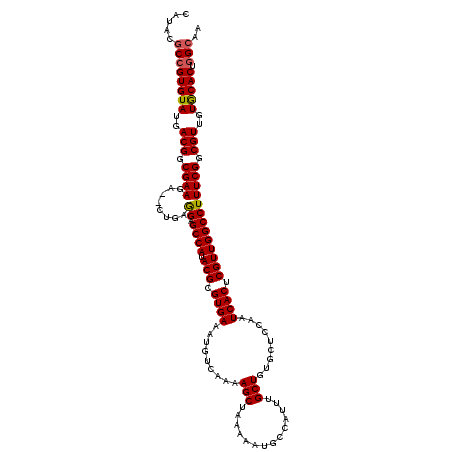
| Location | 731,676 – 731,796 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -26.44 |
| Energy contribution | -30.62 |
| Covariance contribution | 4.18 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 731676 120 + 22407834 UAAGCGAGAACUUCAUGUAGGAGCGUAGGUGCCAAAAUUUUUGCAAGUGCACAACGCCGAAAGGCCAACGAGUGAUUGGAGCACAGCAAAUGGCAUUUUUAGCUUUUGACAUUUCACGCG ...(((.(((....((((((((((..((((((((.....(((((..((((.....(((....)))...(((....)))..)))).)))))))))))))...)))))).))))))).))). ( -42.80) >DroSec_CAF1 3198 120 + 1 UAAGCGGGAACUUCAGUUAGGAGCGUAGGUGCCUCAUUUUUUUCCAGUGCACAACGCCGAAAGGCCAACGAGUGAUUGGAGCACAGCAAAUGGCAUUUUUAGCUUUUGACAUUUCACGCG ...(((((((.....(((((((((..(((((((.........((((((.(((...(((....)))......)))))))))((...))....)))))))...))))))))).)))).))). ( -40.50) >DroSim_CAF1 3204 120 + 1 UAAGCGGGAACUUCAGUUAGGAGCGUAGGUGCCUCAUUUUUUGCCAGUGCACAACGCCGAAAGGCCAACGAGUGAUUGGAGCACAGCAAAUGGCAUUUUUAGCUUUUGACAUUUCACGCG ...(((((((.....(((((((((..(((((((......(((((..((((.....(((....)))...(((....)))..)))).))))).)))))))...))))))))).)))).))). ( -43.80) >DroEre_CAF1 3214 116 + 1 UAAGCAAUAACUUC----AGGAGCGUACUUGGCGAAUUUUUUGCCAGUGCACAACGCCGAAAGGCCAACGAGUGAUUGGAGCACAGCAAAUGGCAUUUUUAGCUUUUGACAUUUCACGCG ...((...((..((----((((((.....((((((.....))))))((((.....(((....)))...(((....)))..)))).((.....)).......))))))))..))....)). ( -34.20) >DroYak_CAF1 3416 119 + 1 UAAGAAAUAACUUUGUGUGGGAGCAUC-CUGUCGAAAUUUUCGCCAGUGUACAACGCCGAAAGGCCAACGAGUGAUUGGAGCACAGCAAAUGGCAUUUUCAGCUUUUGACAUUUCACGCG ..............(((((..(.....-.(((((((......((..((((.(...(((....)))...(((....)))).)))).))....(((.......)))))))))))..))))). ( -30.10) >consensus UAAGCGAGAACUUCAGGUAGGAGCGUAGGUGCCGAAUUUUUUGCCAGUGCACAACGCCGAAAGGCCAACGAGUGAUUGGAGCACAGCAAAUGGCAUUUUUAGCUUUUGACAUUUCACGCG ...(((.(((.......(((((((..((((((((.....(((((..((((.....(((....)))...(((....)))..)))).)))))))))))))...)))))))....))).))). (-26.44 = -30.62 + 4.18)

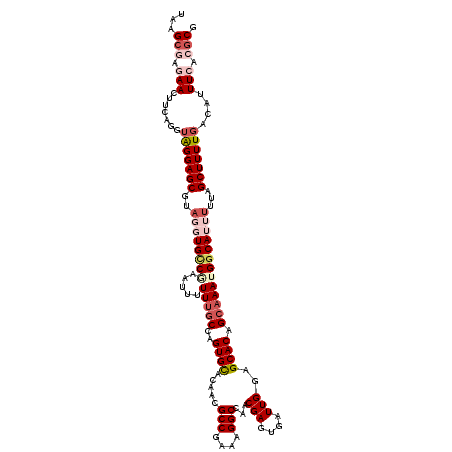
| Location | 731,716 – 731,834 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.13 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -37.91 |
| Energy contribution | -37.99 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 731716 118 - 22407834 CAUACGCCGUGUAUGACGGCGAAGA--CUGAGGAGCCAUACGCGUGAAAUGUCAAAAGCUAAAAAUGCCAUUUGCUGUGCUCCAAUCACUCGUUGGCCUUUCGGCGUUGUGCACUUGCAA .....((.(((((..(((.((((..--....((.((((.(((.((((.........(((..............))).........)))).))))))))))))).)))..)))))..)).. ( -36.51) >DroSec_CAF1 3238 118 - 1 CAUACGCCGUGUAUGACGGCGAAGA--CUGAGGAGCCAUACGCGUGAAAUGUCAAAAGCUAAAAAUGCCAUUUGCUGUGCUCCAAUCACUCGUUGGCCUUUCGGCGUUGUGCACUGGAAA ......(((((((..(((.((((..--....((.((((.(((.((((.........(((..............))).........)))).))))))))))))).)))..))))).))... ( -36.51) >DroSim_CAF1 3244 118 - 1 CAUACGCCGUGUAUGACGGCGAAGA--CUGAGGAGCCAUACGCGUGAAAUGUCAAAAGCUAAAAAUGCCAUUUGCUGUGCUCCAAUCACUCGUUGGCCUUUCGGCGUUGUGCACUGGCAA .....((((((((..(((.((((..--....((.((((.(((.((((.........(((..............))).........)))).))))))))))))).)))..))))).))).. ( -40.61) >DroEre_CAF1 3250 118 - 1 CAUACGCCGUGUAUGACGGCGAAGU--CUGAGGAGCCAUACGCGUGAAAUGUCAAAAGCUAAAAAUGCCAUUUGCUGUGCUCCAAUCACUCGUUGGCCUUUCGGCGUUGUGCACUGGCAA .....((((((((..(((.((((((--(....))((((.(((.((((.........(((..............))).........)))).))))))).))))).)))..))))).))).. ( -40.71) >DroYak_CAF1 3455 120 - 1 CAUACGCCGUGUAUGACGGCGAACUAACUAAAGAGCCAUACGCGUGAAAUGUCAAAAGCUGAAAAUGCCAUUUGCUGUGCUCCAAUCACUCGUUGGCCUUUCGGCGUUGUACACUGGCGA ....(((((((((..(((.((((........((.((((.(((.((((.........(((..............))).........)))).))))))))))))).)))..))))).)))). ( -42.31) >consensus CAUACGCCGUGUAUGACGGCGAAGA__CUGAGGAGCCAUACGCGUGAAAUGUCAAAAGCUAAAAAUGCCAUUUGCUGUGCUCCAAUCACUCGUUGGCCUUUCGGCGUUGUGCACUGGCAA .....((((((((..(((.((((........((.((((.(((.((((.........(((..............))).........)))).))))))))))))).)))..))))).))).. (-37.91 = -37.99 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:16 2006