
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,971,510 – 6,971,655 |
| Length | 145 |
| Max. P | 0.966985 |

| Location | 6,971,510 – 6,971,616 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -21.21 |
| Energy contribution | -22.97 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6971510 106 + 22407834 UGCAA----------AAAAAC-GGUCAGUUUGAGGUUAGGUUGGAGUUUGUUUACUCCACAAGG-GAUAGUAAGCGCUGGCGCAACUCACCUUAAACAGCCAUCCGCUUGGCAAGAAU .....----------......-..((.(((((((((.((..((((((......)))))).....-........(((....)))..)).))))))))).((((......))))..)).. ( -32.20) >DroPse_CAF1 11557 105 + 1 UUCAGACUGUAGUAGAAAAAA-AA--ACAUUUAGGUUAUGUUACGAUUUGUUUACCAUGAAA----------AUCGUGCAAGGAACCAACUUUAAGCAUCCAUCCGUCUGGCCAGAGU .(((((((((...........-..--)))....((((.(((.(((((((...........))----------))))))))...))))..................))))))....... ( -17.42) >DroSec_CAF1 12289 107 + 1 UGCAA---------GAAAAAU-GGUCCGUUUGAGGUUAGGUUGGAGUUUGUUUACUCCGCAAGG-GAGAGUAAACGCUGGCGCAGCUCACCUUAAACAGCCAUCCGCUUGGCAAGAAU (((((---------(....((-(((..(((((((((..(((((..(((((((((((((....))-...))))))))..))).))))).))))))))).)))))...))).)))..... ( -37.10) >DroSim_CAF1 11610 107 + 1 UGCAA---------GAAAAAU-GGUCCGUUUGAGGUUAGGUUGGAGUUUGUUUACUCCGCAAGG-GAGAGUAAACGUUGGCGCAGCUCACCUUAAACAGCCAUCCGCUUGGCAAGAAU (((((---------(....((-(((..(((((((((..(((((..(((((((((((((....))-...))))))))..))).))))).))))))))).)))))...))).)))..... ( -37.10) >DroEre_CAF1 11959 111 + 1 UGCAA-------AAGAAAAAUUGGUAAUUUUGAGGUUAGGUUGAAGUUUGUUUACUCCGCAAGGAGGAAGUAAACGGCAGUGCAGCUCACCUCAAACAGCCAUCAGCUUGGCAAGAAC (((..-------(((......((((...((((((((..(((((..((((((((.((((....)))).)))))))).......))))).))))))))..))))....))).)))..... ( -34.10) >DroYak_CAF1 12105 109 + 1 UGCAG-------AAGAAAAAU-GGUUAGUUUGAGGUUAUGUUAGAGUUUGUUUACUCCGCAAGG-GAAAGUAAACGCUGGUGCAGCUCACCUUAAACGGCCAUCCGCUUGGCAAGAAU (((..-------(((....((-((((.(((((((((.......(((((.(((((((((....))-...)))))))((....))))))))))))))))))))))...))).)))..... ( -36.01) >consensus UGCAA_________GAAAAAU_GGUCAGUUUGAGGUUAGGUUGGAGUUUGUUUACUCCGCAAGG_GAAAGUAAACGCUGGCGCAGCUCACCUUAAACAGCCAUCCGCUUGGCAAGAAU ...........................(((((((((...((((.....((((((((((....))....))))))))......))))..))))))))).((((......))))...... (-21.21 = -22.97 + 1.75)



| Location | 6,971,510 – 6,971,616 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6971510 106 - 22407834 AUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGUUGCGCCAGCGCUUACUAUC-CCUUGUGGAGUAAACAAACUCCAACCUAACCUCAAACUGACC-GUUUUU----------UUGCA .....(((.(((..((((((.((((.((((.(((.(((....)))..)))...-.....((((((......)))))).....)))).)))).).))-)))..)----------))))) ( -29.90) >DroPse_CAF1 11557 105 - 1 ACUCUGGCCAGACGGAUGGAUGCUUAAAGUUGGUUCCUUGCACGAU----------UUUCAUGGUAAACAAAUCGUAACAUAACCUAAAUGU--UU-UUUUUUCUACUACAGUCUGAA ........(((((...((..((((.(((((((((.....)).))))----------)))...))))..))......(((((.......))))--).-..............))))).. ( -16.70) >DroSec_CAF1 12289 107 - 1 AUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGCUGCGCCAGCGUUUACUCUC-CCUUGCGGAGUAAACAAACUCCAACCUAACCUCAAACGGACC-AUUUUUC---------UUGCA .....(((.(((.((((((((((((.((((..((((...))))(((((((((.-......))))))))).............)))).)))))).))-))))..)---------))))) ( -34.80) >DroSim_CAF1 11610 107 - 1 AUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGCUGCGCCAACGUUUACUCUC-CCUUGCGGAGUAAACAAACUCCAACCUAACCUCAAACGGACC-AUUUUUC---------UUGCA .....(((.(((.((((((((((((.(((((((..(((....)))...)))..-......(((((......)))))......)))).)))))).))-))))..)---------))))) ( -31.60) >DroEre_CAF1 11959 111 - 1 GUUCUUGCCAAGCUGAUGGCUGUUUGAGGUGAGCUGCACUGCCGUUUACUUCCUCCUUGCGGAGUAAACAAACUUCAACCUAACCUCAAAAUUACCAAUUUUUCUU-------UUGCA ......((((......))))..(((((((((.((......)))((((((((((.....).))))))))).............))))))))................-------..... ( -26.70) >DroYak_CAF1 12105 109 - 1 AUUCUUGCCAAGCGGAUGGCCGUUUAAGGUGAGCUGCACCAGCGUUUACUUUC-CCUUGCGGAGUAAACAAACUCUAACAUAACCUCAAACUAACC-AUUUUUCUU-------CUGCA ...........((((((((..((((.((((..((((...))))(((((((((.-......))))))))).............)))).))))...))-).......)-------)))). ( -25.91) >consensus AUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGCUGCGCCAGCGUUUACUCUC_CCUUGCGGAGUAAACAAACUCCAACCUAACCUCAAACUGACC_AUUUUUC_________UUGCA ......((((......)))).((((.((((..((((...)))).................(((((......)))))......)))).))))........................... (-14.70 = -14.18 + -0.52)
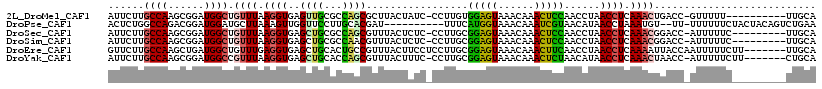
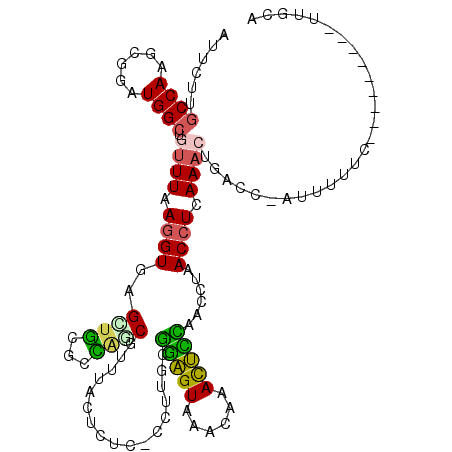
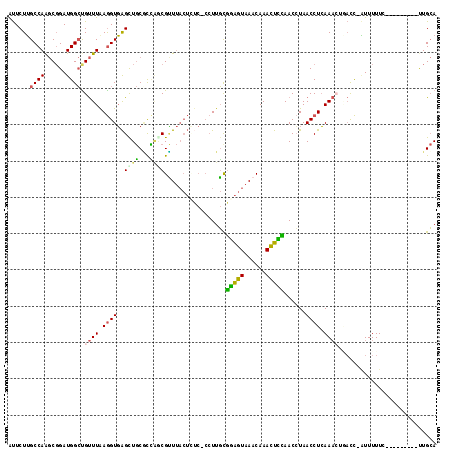
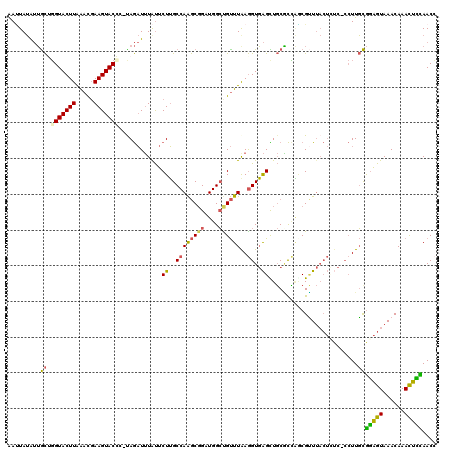
| Location | 6,971,537 – 6,971,655 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -20.34 |
| Energy contribution | -19.52 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6971537 118 - 22407834 AAUUAUAUUACUGGUACUUAAACGAAGUACCC-UAGAUUUAUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGUUGCGCCAGCGCUUACUAUC-CCUUGUGGAGUAAACAAACUCCAACC ............(((((((.....))))))).-...............((((.(((((.((.....))((((((.((....))))))))))))-)))))((((((......))))))... ( -31.90) >DroPse_CAF1 11592 110 - 1 GAUUAAGAAGUUUGUACUUAACUUAAGUACGUACAACUAUACUCUGGCCAGACGGAUGGAUGCUUAAAGUUGGUUCCUUGCACGAU----------UUUCAUGGUAAACAAAUCGUAACA ((((.((..((.((((((((...)))))))).))..))....((((......))))((..((((.(((((((((.....)).))))----------)))...))))..))))))...... ( -21.00) >DroSec_CAF1 12317 118 - 1 AAUUAUAUUGCGGGUACUUAAACGAAGUACCG-CAGAUUUAUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGCUGCGCCAGCGUUUACUCUC-CCUUGCGGAGUAAACAAACUCCAACC .........((.(((((((.....)))))))(-(((.......((..((((((((....))))))..))..))))))....))(((((((((.-......)))))))))........... ( -35.61) >DroSim_CAF1 11638 118 - 1 AAUUAUAUUGCGGGUACUUAAACGAAGUACCG-UAGAUUUAUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGCUGCGCCAACGUUUACUCUC-CCUUGCGGAGUAAACAAACUCCAACC .........((.(((((((.....)))))))(-((((.....)).)))...))((.(((..((((..((((.....))))...(((((((((.-......))))))))))))).))).)) ( -33.30) >DroEre_CAF1 11990 119 - 1 AAGUAUAUUGCUGGUACUUAGACGAAGUACGC-UAGAUUUGUUCUUGCCAAGCUGAUGGCUGUUUGAGGUGAGCUGCACUGCCGUUUACUUCCUCCUUGCGGAGUAAACAAACUUCAACC .........(((.((((((.....))))))((-.(((.....))).))..)))(((.(..((((((((((((((.((...)).)))))))))((((....)))).)))))..).)))... ( -32.40) >DroYak_CAF1 12135 118 - 1 AACUAUAUUGCUGGUACUUGAACGAAGUACGC-UAGAUUUAUUCUUGCCAAGCGGAUGGCCGUUUAAGGUGAGCUGCACCAGCGUUUACUUUC-CCUUGCGGAGUAAACAAACUCUAACA .........((((((((((.....))))))((-.(((.....))).))...((((.(.(((......))).).))))..))))(((((((((.-......)))))))))........... ( -33.10) >consensus AAUUAUAUUGCUGGUACUUAAACGAAGUACCC_UAGAUUUAUUCUUGCCAAGCGGAUGGCUGUUUAAGGUGAGCUGCGCCAGCGUUUACUCUC_CCUUGCGGAGUAAACAAACUCCAACC .........((.(((((((.....)))))))............((..((((((((....))))))..))..)).........................))(((((......))))).... (-20.34 = -19.52 + -0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:55 2006