| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,970,460 – 6,970,565 |
| Length | 105 |
| Max. P | 0.781003 |

| Location | 6,970,460 – 6,970,565 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -20.49 |
| Energy contribution | -22.60 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
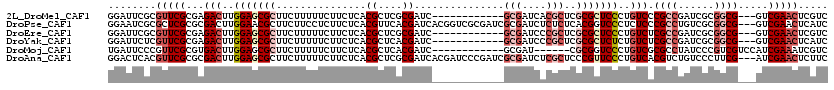
>2L_DroMel_CAF1 6970460 105 + 22407834 GGAUUCGCGUUCGCGAGACUUGGAGCGCUUCUUUUUCUUCUCACGCUCGCGAUC------------GCGAUCACGCUCGCGCUCCCUGUCCCGCCGAUCGCGGCG---GUCGAACUCGUC .....((.(((((...(((..(((((((...............((....))...------------(((....)))..)))))))..)))((((((....)))))---).))))).)).. ( -40.90) >DroPse_CAF1 10496 117 + 1 GGAAUCGCGCUCGCGCGACUUGGAACGCUUCUUCCUCUUCUCACGUUCACGAUCACGGUCGCGAUCGCGAUCUCUCUCACGGUCCCUCUCCCGCCUGUCGCGGCG---GUCGAACUCAUC (..((((((....(((((((.((((......))))........((....)).....)))))))..))))))..)............((..(((((......))))---)..))....... ( -36.20) >DroEre_CAF1 10911 105 + 1 GGAUUCGCGUUCGCGAGACUUGGAGCGCUUCUUUUUCUUCUCACGCUCGCGAUC------------GCGAUCCCGCUCGCGCUCCCUGUCUCGCCGAUCGCGGCG---GUCGAACUCGUC .((((((((.(((((((((..(((((((...............((....))...------------(((....)))..)))))))..)))))).))).)))))..---)))......... ( -43.70) >DroYak_CAF1 11056 105 + 1 GGAUUCUCGUUCGCGAGACUUGGAGCGCUUCUUUUUCUUCUCACGCUCACGAUC------------GCGAUCCCGCUCGCGCUCUCUGUCUCGCCGAUCGCGGCG---GUCGAACUCAUC ........(((((((((((..(((((((...............((....))...------------(((....)))..)))))))..))))))).(((((...))---)))))))..... ( -38.90) >DroMoj_CAF1 11801 102 + 1 UGAUUCCCGUUCGCGUGACUUGGAGCGCUUCUUUUUCUUCUCACGCUCACGAUC------------GCGAU------CGCGGUCCCUGUCGCGCCUAUCCCGUCGUCCAUCGAAAUCGUC .((((.......(((((((...(((((................)))))..((((------------((...------.))))))...)))))))........(((.....)))))))... ( -29.29) >DroAna_CAF1 8238 117 + 1 GGACUCACGUUCGCGCGACUUGGAGCGCUUCUUUUUCUUCUCACGCUCGCGAUCACGAUCCCGAUCGCGAUCUCGCUCCCGUUCCCUGUCACGUCUGUCCCUUCG---AUCGAACUCUUC ((((..((((.((.(.(((..(((((((................))((((((((........))))))))....))))).))).).))..))))..)))).....---............ ( -32.79) >consensus GGAUUCGCGUUCGCGAGACUUGGAGCGCUUCUUUUUCUUCUCACGCUCACGAUC____________GCGAUCCCGCUCGCGCUCCCUGUCCCGCCGAUCGCGGCG___GUCGAACUCGUC ........(((((...(((..(((((((...............((....))...............(((....)))..)))))))..))).((((......)))).....)))))..... (-20.49 = -22.60 + 2.11)

| Location | 6,970,460 – 6,970,565 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -24.40 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6970460 105 - 22407834 GACGAGUUCGAC---CGCCGCGAUCGGCGGGACAGGGAGCGCGAGCGUGAUCGC------------GAUCGCGAGCGUGAGAAGAAAAAGAAGCGCUCCAAGUCUCGCGAACGCGAAUCC ..((.(((((.(---(((((....))))))(((..(((((((...((((((...------------.))))))..(....)...........)))))))..)))...))))).))..... ( -47.60) >DroPse_CAF1 10496 117 - 1 GAUGAGUUCGAC---CGCCGCGACAGGCGGGAGAGGGACCGUGAGAGAGAUCGCGAUCGCGACCGUGAUCGUGAACGUGAGAAGAGGAAGAAGCGUUCCAAGUCGCGCGAGCGCGAUUCC .......((..(---((((......)))))..))((((.((((.......((((((((((....)))))))))).((((.((...((((......))))...)).))))..)))).)))) ( -47.70) >DroEre_CAF1 10911 105 - 1 GACGAGUUCGAC---CGCCGCGAUCGGCGAGACAGGGAGCGCGAGCGGGAUCGC------------GAUCGCGAGCGUGAGAAGAAAAAGAAGCGCUCCAAGUCUCGCGAACGCGAAUCC ..((.(((((..---.((((....))))(((((..(((((((..(((....)))------------..((((....))))............)))))))..))))).))))).))..... ( -46.40) >DroYak_CAF1 11056 105 - 1 GAUGAGUUCGAC---CGCCGCGAUCGGCGAGACAGAGAGCGCGAGCGGGAUCGC------------GAUCGUGAGCGUGAGAAGAAAAAGAAGCGCUCCAAGUCUCGCGAACGAGAAUCC (((..(((((..---.((((....))))(((((...((.(((((......))))------------).))(.((((((..............)))))))..))))).)))))....))). ( -39.54) >DroMoj_CAF1 11801 102 - 1 GACGAUUUCGAUGGACGACGGGAUAGGCGCGACAGGGACCGCG------AUCGC------------GAUCGUGAGCGUGAGAAGAAAAAGAAGCGCUCCAAGUCACGCGAACGGGAAUCA ...(((((((((..((((((.(((...((((........))))------))).)------------).))))((((((..............))))))...))).((....)))))))). ( -29.24) >DroAna_CAF1 8238 117 - 1 GAAGAGUUCGAU---CGAAGGGACAGACGUGACAGGGAACGGGAGCGAGAUCGCGAUCGGGAUCGUGAUCGCGAGCGUGAGAAGAAAAAGAAGCGCUCCAAGUCGCGCGAACGUGAGUCC .....(((((.(---(....)).....((((((.(....).((((((...(((((((((......))))))))).(....)............))))))..)))))))))))........ ( -42.70) >consensus GACGAGUUCGAC___CGCCGCGAUAGGCGAGACAGGGAGCGCGAGCGAGAUCGC____________GAUCGCGAGCGUGAGAAGAAAAAGAAGCGCUCCAAGUCUCGCGAACGCGAAUCC .....(((((......(((......)))(((((.......((((......))))................(.((((((..............)))))))..))))).)))))........ (-24.40 = -25.16 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:52 2006