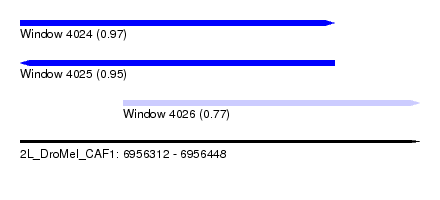
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,956,312 – 6,956,448 |
| Length | 136 |
| Max. P | 0.972570 |

| Location | 6,956,312 – 6,956,419 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.58 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -10.68 |
| Energy contribution | -11.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6956312 107 + 22407834 UAAAA-CCUCCUUAACUACACU-UU---AUAAACACAACACAUGUUUGCC------GAUCUAGCCAAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA-UUAGUAAAAAGUUUUAA (((((-(.......((((....-.(---(((....((((((..(((((.(------......).)))))((((((....))))))....).))))).))))-.)))).....)))))). ( -19.50) >DroSec_CAF1 5779 104 + 1 CAAAA-CCUCCUUAACAC-------------AACACAAUAAACCUAUGCCAAUUGUGAUCUAGCCCAACACGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA-UUAGUAAAAAGUAUUAA .....-......(((.((-------------..((((((............))))))...((((..(((((((((..........))))).))))))))..-..........)).))). ( -14.60) >DroSim_CAF1 5782 104 + 1 CAAAA-CCUCCUUAACAC-------------AACACAAUAAACCUAUGUCAAUUGUGAUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA-UUAGUAAAAAGUAUUAA .....-......(((.((-------------..((((((..((....))..))))))...((((.....((((((....))))))....))))((((((..-.))))))...)).))). ( -18.60) >DroEre_CAF1 5836 102 + 1 UAAACAGAGAGGUAUUUGGGUAGUUUGGUUAAUCA-------UCUA---CUU------UCUUGACCCAUGCGAAUAAAAAUCCGCAUCCGCUGUAGCUAUU-UUAGUUAUAGGUAUUAA ........(.(((....((((((..(((....)))-------.)))---)))------.....))))(((((..........)))))...(((((((((..-.)))))))))....... ( -20.40) >DroYak_CAF1 5795 102 + 1 UAACAAAAGCGUUAAUAAGGUU-UUUGGUUAAACA-------CCUA---CAU------UUACGACCCAUGCGGAUAAAAAUCCGCAUCCGCUGUAUCUAUAUAUAGUGAUAAGUAUUAG ...(((((((.(.....).)))-))))........-------....---...------.(((.....((((((((....)))))))).((((((((....))))))))....))).... ( -23.70) >consensus UAAAA_CCUCCUUAACAA_____UU____UAAACACAA_A_ACCUAUG_CAA____GAUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA_UUAGUAAAAAGUAUUAA .....................................................................((((((....))))))....(((..(((((....)))))...)))..... (-10.68 = -11.08 + 0.40)


| Location | 6,956,312 – 6,956,419 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.58 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.40 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6956312 107 - 22407834 UUAAAACUUUUUACUAA-UAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUUGGCUAGAUC------GGCAAACAUGUGUUGUGUUUAU---AA-AGUGUAGUUAAGGAGG-UUUUA .(((((((((((((((.-(((((..((((((.((((((((....))))))(((((.(......------).))))))).))))))..))))---).-....))))..))))))-))))) ( -32.30) >DroSec_CAF1 5779 104 - 1 UUAAUACUUUUUACUAA-UAUAGCAACAGCGGAUGCGGAUUUUUAUCCGUGUUGGGCUAGAUCACAAUUGGCAUAGGUUUAUUGUGUU-------------GUGUUAAGGAGG-UUUUG .(((.(((((((..(((-((((((...(((.(((((((((....))))))))).).)).....(((((..((....))..))))))))-------------))))))))))))-).))) ( -27.50) >DroSim_CAF1 5782 104 - 1 UUAAUACUUUUUACUAA-UAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUGGGCUAGAUCACAAUUGACAUAGGUUUAUUGUGUU-------------GUGUUAAGGAGG-UUUUG .(((.(((((((..(((-((((((...(((.(((((((((....))))))))).).)).....(((((.(((....))).))))))))-------------))))))))))))-).))) ( -30.30) >DroEre_CAF1 5836 102 - 1 UUAAUACCUAUAACUAA-AAUAGCUACAGCGGAUGCGGAUUUUUAUUCGCAUGGGUCAAGA------AAG---UAGA-------UGAUUAACCAAACUACCCAAAUACCUCUCUGUUUA ................(-(((((....((.((((((((((....))))))))((((.....------.((---(...-------((......)).))))))).....)).)))))))). ( -17.30) >DroYak_CAF1 5795 102 - 1 CUAAUACUUAUCACUAUAUAUAGAUACAGCGGAUGCGGAUUUUUAUCCGCAUGGGUCGUAA------AUG---UAGG-------UGUUUAACCAAA-AACCUUAUUAACGCUUUUGUUA .(((((............((((..(((.((..((((((((....))))))))..)).))).------.))---))((-------(.....)))...-.....)))))............ ( -21.40) >consensus UUAAUACUUUUUACUAA_UAUAGCAACAGCGGAUGCGGAUUUUUAUCCGCGUUGGGCUAGAUC____AUG_CAUAGGU_U_UUGUGUUUA____AA_____GUGUUAAGGAGG_UUUUA .............................(.(((((((((....))))))))).)................................................................ (-11.56 = -11.40 + -0.16)


| Location | 6,956,347 – 6,956,448 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.14 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6956347 101 + 22407834 CAUGUUUGCC------GAUCUAGCCAAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA-UUAGUAAAAAGUUUUAAAUAAGAGAAGGCAUAAAUUACUUUAUGAA ......((((------..((((((.....((((((....))))))....))).((((((..-.)))))).............)))...))))................ ( -21.30) >DroSec_CAF1 5805 107 + 1 AACCUAUGCCAAUUGUGAUCUAGCCCAACACGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA-UUAGUAAAAAGUAUUAAAUAGGAGAAUGAUUAAAUUACUAUUGGAA ........(((((.(((((.(((..((((((((((..........))))).)))))((((.-((((((.....))))))))))........))).))))).))))).. ( -21.80) >DroSim_CAF1 5808 107 + 1 AACCUAUGUCAAUUGUGAUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA-UUAGUAAAAAGUAUUAAAUAAGAGAAGGAUUAAAUUACUUUUGGAA ........((((..(((((.((..((...((((((....))))))....(((.((((((..-.))))))..)))..............))..)).)))))..)))).. ( -23.30) >DroEre_CAF1 5871 96 + 1 --UCUA---CUU------UCUUGACCCAUGCGAAUAAAAAUCCGCAUCCGCUGUAGCUAUU-UUAGUUAUAGGUAUUAAAUAAGAAGAUGUUUCGAUCACUUUUGGAA --(((.---(((------..((((..((((((..........)))))...(((((((((..-.))))))))))..))))..))).)))..((((((......)))))) ( -17.70) >DroYak_CAF1 5829 97 + 1 --CCUA---CAU------UUACGACCCAUGCGGAUAAAAAUCCGCAUCCGCUGUAUCUAUAUAUAGUGAUAAGUAUUAGAUAAGAAAAUGUACAAAUCACUUUUGGAA --..((---(((------((.(.....((((((((....)))))))).((((((((....))))))))...............).)))))))((((.....))))... ( -23.20) >consensus _ACCUAUG_CAA____GAUCUAGCCCAACGCGGAUAAAAAUCCGCAUCCGCUGUUGCUAUA_UUAGUAAAAAGUAUUAAAUAAGAGAAUGAUUAAAUUACUUUUGGAA ........................((...((((((....))))))....(((..(((((....)))))...)))..............................)).. (-12.06 = -12.14 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:44 2006