| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,940,874 – 6,940,975 |
| Length | 101 |
| Max. P | 0.994902 |

| Location | 6,940,874 – 6,940,975 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -18.27 |
| Energy contribution | -17.67 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
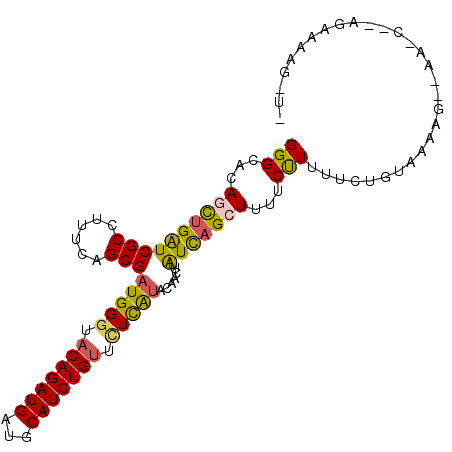
>2L_DroMel_CAF1 6940874 101 + 22407834 GGGCACAACUGGUCGCCUUUCAGCGAUGGGUACAGAUGAUGCAUCUGUUCUCGUACAACUUAUCAGCUUUUCUUUUUCUGCAAGAGUUGAAG--UGUGAAG-UC ..((((.(((.(((((......))))).)))(((((((...)))))))..............((((((((((.......).))))))))).)--)))....-.. ( -28.60) >DroVir_CAF1 10081 97 + 1 GGGCAGAGCUGAUCGCCUUUCAGCGAUGGGUACAGAUGAUGCAUCUGUUCUUAAAUGAAUAAUCAGCUUUGCCUUCUCUGUAAGUU--CA-GG-AAAAAAA--- .(((((((((((((((......))))((((.(((((((...))))))).)))).........)))))))))))(((.(((......--))-))-)).....--- ( -36.90) >DroGri_CAF1 26259 100 + 1 GGGCAGAGCUGAUCGCCUUUCAGCGAUGGGUACAGAUGAUGCAUCUGUCUUUAUAUAAAUUGUCAGCUUUGCCUUUUCUGUAAAUU--AU-CG-AAACAAAUUU ((((((((((((((((......)))(((((.(((((((...))))))).))))).......)))))))))))))............--..-..-.......... ( -32.00) >DroWil_CAF1 9392 100 + 1 GGGCACAAUUGAUCGCCCUUCAGCGAUGGGUACAGAUGAUGCAUCUGUUUUUAUGAAAUUGAUCAGCUUUUCUUUUUCUGUGAAAGACAAAA--AGAAAAG-A- ........((((((((((.........))))(((((((...)))))))............))))))(((((((((((((.....)))..)))--)))))))-.- ( -27.10) >DroYak_CAF1 8446 103 + 1 GGGCACAGCUGGUCGCCUUUCAGCGAUGGGUACAGAUGAUGCAUCUGUUCUCGUACAACUUAUCCGCUUUUCUUUUUCUGCAAGAGUUGAACGUUGUGAAG-UU ...((((((..(((((......)))))(((.(((((((...))))))).)))((.((((((....((............))..)))))).))))))))...-.. ( -30.30) >DroAna_CAF1 7753 84 + 1 GGGCAGAGCUGAUCGCCUUUCAGCGAUGGAUACAGAUGAUGCAUCUGAUCUCGUACAACUUGUUAGCUUUUCUUUUUCUGUAAA-------------------- ..((((((((((.......))))).((((((.((((((...))))))))).)))......................)))))...-------------------- ( -21.80) >consensus GGGCACAGCUGAUCGCCUUUCAGCGAUGGGUACAGAUGAUGCAUCUGUUCUCAUACAACUUAUCAGCUUUUCUUUUUCUGUAAAAG__AA_C__AGAAAAG_U_ (((...((((((((((......)))(((((.(((((((...))))))).))))).......)))))))...))).............................. (-18.27 = -17.67 + -0.61)

| Location | 6,940,874 – 6,940,975 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
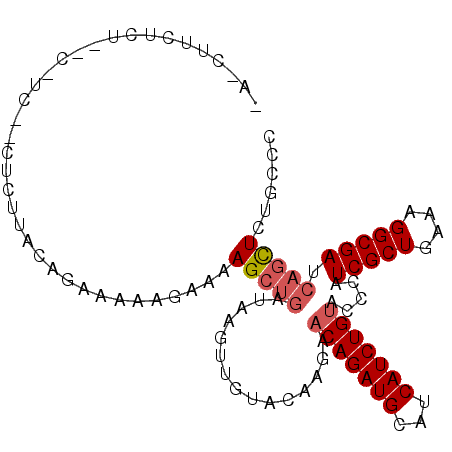
>2L_DroMel_CAF1 6940874 101 - 22407834 GA-CUUCACA--CUUCAACUCUUGCAGAAAAAGAAAAGCUGAUAAGUUGUACGAGAACAGAUGCAUCAUCUGUACCCAUCGCUGAAAGGCGACCAGUUGUGCCC ((-(((..((--(((....((((.......)))).))).))..)))))((((((..(((((((...))))))).....(((((....)))))....)))))).. ( -23.70) >DroVir_CAF1 10081 97 - 1 ---UUUUUUU-CC-UG--AACUUACAGAGAAGGCAAAGCUGAUUAUUCAUUUAAGAACAGAUGCAUCAUCUGUACCCAUCGCUGAAAGGCGAUCAGCUCUGCCC ---.....((-((-((--......))).)))((((.((((((..............(((((((...))))))).....(((((....))))))))))).)))). ( -33.90) >DroGri_CAF1 26259 100 - 1 AAAUUUGUUU-CG-AU--AAUUUACAGAAAAGGCAAAGCUGACAAUUUAUAUAAAGACAGAUGCAUCAUCUGUACCCAUCGCUGAAAGGCGAUCAGCUCUGCCC ...(((((..-..-..--.....)))))...((((.((((((..............(((((((...))))))).....(((((....))))))))))).)))). ( -31.70) >DroWil_CAF1 9392 100 - 1 -U-CUUUUCU--UUUUGUCUUUCACAGAAAAAGAAAAGCUGAUCAAUUUCAUAAAAACAGAUGCAUCAUCUGUACCCAUCGCUGAAGGGCGAUCAAUUGUGCCC -.-(((((((--((((.(.......).))))))))))).(((((............(((((((...))))))).(((.((...)).))).)))))......... ( -23.70) >DroYak_CAF1 8446 103 - 1 AA-CUUCACAACGUUCAACUCUUGCAGAAAAAGAAAAGCGGAUAAGUUGUACGAGAACAGAUGCAUCAUCUGUACCCAUCGCUGAAAGGCGACCAGCUGUGCCC ..-...((((.(((.(((((.((((............))))...))))).)))...(((((((...))))))).....(((((....))))).....))))... ( -25.30) >DroAna_CAF1 7753 84 - 1 --------------------UUUACAGAAAAAGAAAAGCUAACAAGUUGUACGAGAUCAGAUGCAUCAUCUGUAUCCAUCGCUGAAAGGCGAUCAGCUCUGCCC --------------------....((((...((.....)).....((((...(.(((((((((...)))))).))))((((((....))))))))))))))... ( -20.50) >consensus _A_CUUCUCU__C_UC__CUCUUACAGAAAAAGAAAAGCUGAUAAGUUGUACAAGAACAGAUGCAUCAUCUGUACCCAUCGCUGAAAGGCGAUCAGCUCUGCCC ....................................(((((...............(((((((...))))))).....(((((....))))).)))))...... (-16.95 = -17.40 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:41 2006