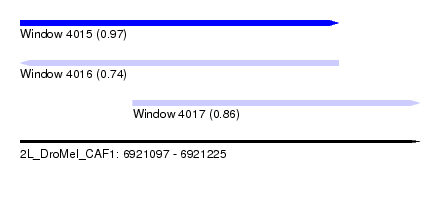
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,921,097 – 6,921,225 |
| Length | 128 |
| Max. P | 0.965526 |

| Location | 6,921,097 – 6,921,199 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -13.18 |
| Energy contribution | -14.57 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.27 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
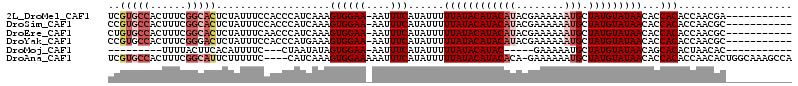
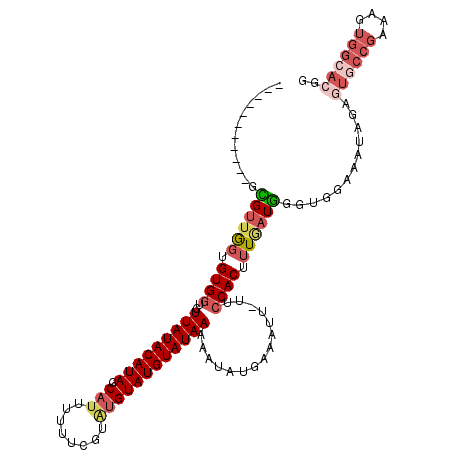
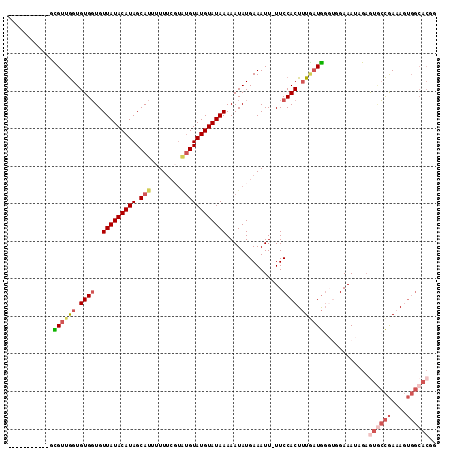
>2L_DroMel_CAF1 6921097 102 + 22407834 UCGUGCCACUUUCGGCACUCUAUUUCCACCCAUCAAAGUGGAA-AAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGA----------- ..(((((......)))))....(((((((........))))))-)...........((((((((((((........))).)))))))))..............----------- ( -24.40) >DroSim_CAF1 6493 102 + 1 CCGUGCCACUUUCGGCACUCUAUUUCCACCCAUCAAAGUGGAA-AAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGC----------- ..(((((......)))))....(((((((........))))))-)...........((((((((((((........))).)))))))))..............----------- ( -24.40) >DroEre_CAF1 7546 102 + 1 CUGUGCCACUUUCGGCACUCUAUUUCAACCCAUCAAAGUGGAA-AAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGC----------- ..(((((......)))))...................((((((-(((.....)))))(((((((((((........))).))))))))...))))........----------- ( -21.20) >DroYak_CAF1 6216 102 + 1 CCGUGCCACUUUCGGGACUCUAUUUCCACCCAUGAAAGUGGAA-AAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGC----------- ..((((((((((((((............)))..))))))))..-............((((((((((((........))).)))))))))...)))........----------- ( -23.80) >DroMoj_CAF1 6085 85 + 1 ---------UUUUACUUCACAUUUUC---CUAAUAUAGUGGAA-AAUUUCAUAUUUUUAUACAUAC-----GAAAAAUGCUAUGUAUAACAGCACACUAACAC----------- ---------...........((((((---(.........))))-))).........((((((((((-----(.....)).)))))))))..............----------- ( -10.50) >DroAna_CAF1 6051 109 + 1 UCGUGCCACUUUCGGCAUUCUUUUUC----CAUCAAAGUGGAAAAAUUUCAUAUUUUUAUACAUACACA-GAAAAAAUGCUAUGUAUAACACCACACCAACACUGGCAAAGCCA ..(((((......)))))..((((((----(((....)))))))))..........(((((((((..((-.......)).)))))))))...............(((...))). ( -22.30) >consensus CCGUGCCACUUUCGGCACUCUAUUUCCACCCAUCAAAGUGGAA_AAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGC___________ ..(((((......)))))...................((((((....)))......((((((((((((........))).)))))))))...)))................... (-13.18 = -14.57 + 1.39)
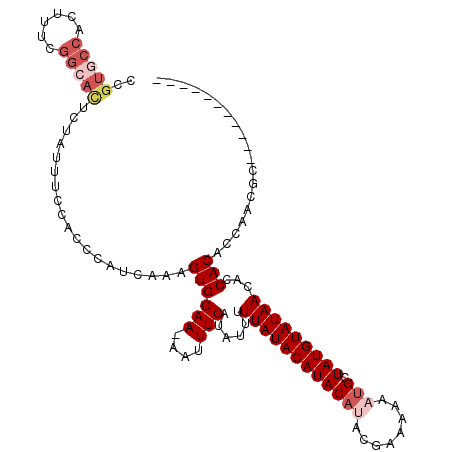
| Location | 6,921,097 – 6,921,199 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -16.03 |
| Energy contribution | -17.65 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6921097 102 - 22407834 -----------UCGUUGGUGUGGUGUUAUACAUAGCAUUUUUUCGUAUGUAUGUAUAAAAAUAUGAAAUU-UUCCACUUUGAUGGGUGGAAAUAGAGUGCCGAAAGUGGCACGA -----------...((.((((....(((((((((.(((........))))))))))))..)))).))..(-((((((((....)))))))))....((((((....)))))).. ( -30.40) >DroSim_CAF1 6493 102 - 1 -----------GCGUUGGUGUGGUGUUAUACAUAGCAUUUUUUCGUAUGUAUGUAUAAAAAUAUGAAAUU-UUCCACUUUGAUGGGUGGAAAUAGAGUGCCGAAAGUGGCACGG -----------...((.((((....(((((((((.(((........))))))))))))..)))).))..(-((((((((....)))))))))....((((((....)))))).. ( -30.40) >DroEre_CAF1 7546 102 - 1 -----------GCGUUGGUGUGGUGUUAUACAUAGCAUUUUUUCGUAUGUAUGUAUAAAAAUAUGAAAUU-UUCCACUUUGAUGGGUUGAAAUAGAGUGCCGAAAGUGGCACAG -----------.(((..(.((((..(((((((((.(((........))))))))))))............-..)))).)..)))............((((((....)))))).. ( -25.89) >DroYak_CAF1 6216 102 - 1 -----------GCGUUGGUGUGGUGUUAUACAUAGCAUUUUUUCGUAUGUAUGUAUAAAAAUAUGAAAUU-UUCCACUUUCAUGGGUGGAAAUAGAGUCCCGAAAGUGGCACGG -----------...((.((((....(((((((((.(((........))))))))))))..)))).))..(-((((((((....)))))))))....(((((....).)).)).. ( -26.50) >DroMoj_CAF1 6085 85 - 1 -----------GUGUUAGUGUGCUGUUAUACAUAGCAUUUUUC-----GUAUGUAUAAAAAUAUGAAAUU-UUCCACUAUAUUAG---GAAAAUGUGAAGUAAAA--------- -----------(((((((((((....))))).))))))..(((-----((((........)))))))(((-((((.........)---))))))...........--------- ( -15.90) >DroAna_CAF1 6051 109 - 1 UGGCUUUGCCAGUGUUGGUGUGGUGUUAUACAUAGCAUUUUUUC-UGUGUAUGUAUAAAAAUAUGAAAUUUUUCCACUUUGAUG----GAAAAAGAAUGCCGAAAGUGGCACGA ......(((((.(.(((((((....(((((((((.(((......-.))))))))))))..........((((((((......))----))))))..))))))).).)))))... ( -30.60) >consensus ___________GCGUUGGUGUGGUGUUAUACAUAGCAUUUUUUCGUAUGUAUGUAUAAAAAUAUGAAAUU_UUCCACUUUGAUGGGUGGAAAUAGAGUGCCGAAAGUGGCACGG ............((((((.((((..(((((((((.(((........))))))))))))...............)))).))))))............((((((....)))))).. (-16.03 = -17.65 + 1.62)

| Location | 6,921,133 – 6,921,225 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -13.75 |
| Consensus MFE | -8.64 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
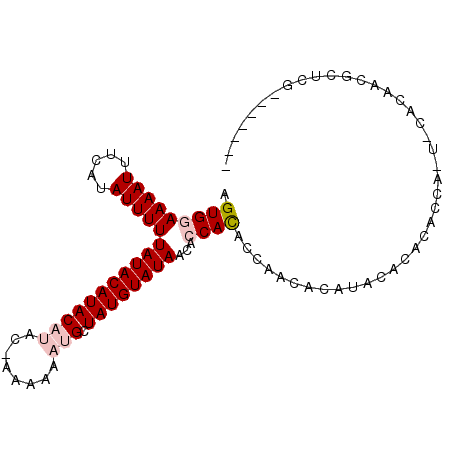
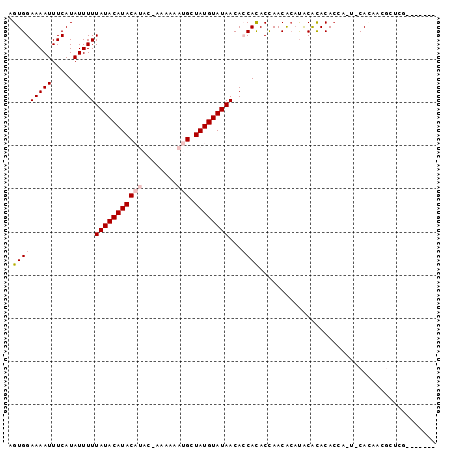
>2L_DroMel_CAF1 6921133 92 + 22407834 AGUGGAAAAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGAUCACGCACACCAUUGCACAGCGCUCG------- .(((((((((.....)))))(((((((((((........))).))))))))...)))).....(((...(((.............))).)))------- ( -15.32) >DroVir_CAF1 6406 81 + 1 AGUGGAAAAUUUCAUAUUUUUAUACAUAC-----AAAAAAUGCUAUGUAUAACAGCAUACUAACACAUACAUACA------CACAAGAUACA------- .(((((((((.....))))))((((((((-----(.....)).))))))).............))).........------...........------- ( -9.00) >DroPse_CAF1 5813 98 + 1 AGUGGAAAAUUUCAUAUUUUUAUACAUACAUAC-AAAAAAUGCUAUGUAUAACACCACACCAACACAUACACACACGAGUACAAAUCGGUCGGUUCGGU .(((((((((.....)))))(((((((((((..-.....))).))))))))...))))((((((........((.(((.......)))))..))).))) ( -15.90) >DroSec_CAF1 6098 92 + 1 AGUGGAAAAUUUCAUAUUUUUAUACAUACAUACGAAAAAAUGCUAUGUAUAACACCACACCAACGCAUACGCACACCAUUGCAUAGAGCUCG------- .(((((((((.....)))))(((((((((((........))).))))))))...))))......((.((.(((......))).))..))...------- ( -16.50) >DroMoj_CAF1 6109 83 + 1 AGUGGAAAAUUUCAUAUUUUUAUACAUAC-----GAAAAAUGCUAUGUAUAACAGCACACUAACACAUGCAUACACA----CACAAGACACA------- .(((...............((((((((((-----(.....)).)))))))))..(((..........)))...))).----...........------- ( -9.90) >DroPer_CAF1 5813 98 + 1 AGUGGAAAAUUUCAUAUUUUUAUACAUACAUAC-AAAAAAUGCUAUGUAUAACACCACACCAACACAUACACACACGAGUACAAAUCGGUCGGUUCGGU .(((((((((.....)))))(((((((((((..-.....))).))))))))...))))((((((........((.(((.......)))))..))).))) ( -15.90) >consensus AGUGGAAAAUUUCAUAUUUUUAUACAUACAUAC_AAAAAAUGCUAUGUAUAACACCACACCAACACAUACACACACCA_U_CACAACGCUCG_______ .(((((((((.....)))))(((((((((((........))).))))))))...))))......................................... ( -8.64 = -9.50 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:36 2006