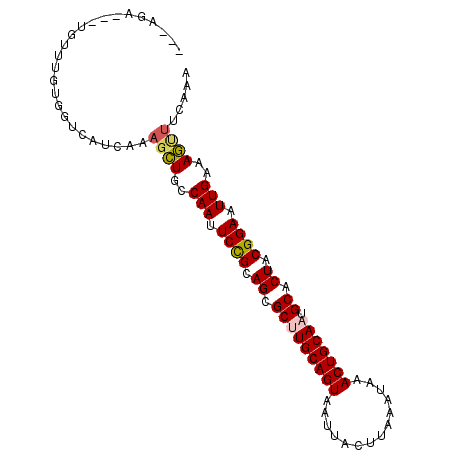
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,902,066 – 6,902,159 |
| Length | 93 |
| Max. P | 0.994500 |

| Location | 6,902,066 – 6,902,159 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.10 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -21.97 |
| Energy contribution | -20.97 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6902066 93 + 22407834 UUUGAAGUUUUCAAUUCCGUAGUGCAUUGCAGUGUGUUUCAUUAUUUACUGCAAGCGCUGCGGAUUUGGCAACUUUGACGACCUUCAUCA---UCU--- .(..(((((.((((.((((((((((.((((((((.((......)).)))))))))))))))))).)))).)))))..)............---...--- ( -39.00) >DroVir_CAF1 134764 96 + 1 AGCGAAACUCUCAAUUCCGUAGUGCAUUGCAGUUGAUUUUAGGAAUUACUGCAAGCGCUGCAGAAUUGGCAGCUCCGCUGACCGCAGACAACGUAU--- .(((......(((((((.(((((((.(((((((.(((((...))))))))))))))))))).)))))))((((...))))..)))...........--- ( -35.90) >DroGri_CAF1 121368 96 + 1 AAUGUAACUUUCAAUUCCGUAGUGCAUUGCAGUUGAUUUAAGCAACUACUGCAUGCCCUGCAGAAUUGGCAGCUCUGUUGACCACAUACAGCAAAU--- .......((.(((((((.((((.((((.(((((..............))))))))).)))).))))))).))...(((((........)))))...--- ( -26.74) >DroEre_CAF1 100712 96 + 1 UUUGAAGUUUUCAAUUCCGUAGUGCAUUGCAGUGUCUUUACUUAUUUACUGCAAGCGCUGCGGAUUUGGCAACUUUGACGACCUUAAUCA---UCUGGA .(..(((((.((((.((((((((((.((((((((............)))))))))))))))))).)))).)))))..)...((.......---...)). ( -38.70) >DroMoj_CAF1 147675 96 + 1 UGCGUAACUUUCAAUUCCGUAGUGCAUUGCAGUUGAUUUAAGGAAUUACUGCAAGCGCUGCAGAAUUGGCAGCUCCGUUGACCACAGACAGCAUAU--- (((....((.(((((((.(((((((.(((((((.(((((...))))))))))))))))))).))))))).))....(((.......))).)))...--- ( -32.70) >DroAna_CAF1 94286 93 + 1 UUCGAGACUUUCAAUUCCGUAGUGCAUUGCAGUGUUUUUAAACUACAACUGCAUGCUCUGCGGACUUGGCAGAUUUGAAGACCAGGAGCA---ACU--- ((((((.((.((((.(((((((.((((.(((((((.........)).))))))))).))))))).)))).)).))))))...........---...--- ( -32.60) >consensus UUCGAAACUUUCAAUUCCGUAGUGCAUUGCAGUGGAUUUAAGCAAUUACUGCAAGCGCUGCAGAAUUGGCAGCUCUGAUGACCACAAACA___UAU___ .(((.(.((.((((.((.(((((((.(((((((..............)))))))))))))).)).)))).)).).)))..................... (-21.97 = -20.97 + -1.00)

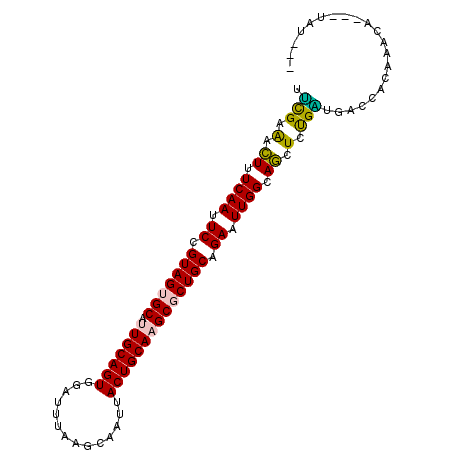

| Location | 6,902,066 – 6,902,159 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.10 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6902066 93 - 22407834 ---AGA---UGAUGAAGGUCGUCAAAGUUGCCAAAUCCGCAGCGCUUGCAGUAAAUAAUGAAACACACUGCAAUGCACUACGGAAUUGAAAACUUCAAA ---.((---((((....)))))).(((((..(((.((((.((.(((((((((..............))))))).)).)).)))).)))..))))).... ( -29.54) >DroVir_CAF1 134764 96 - 1 ---AUACGUUGUCUGCGGUCAGCGGAGCUGCCAAUUCUGCAGCGCUUGCAGUAAUUCCUAAAAUCAACUGCAAUGCACUACGGAAUUGAGAGUUUCGCU ---...(((.....)))...(((((((((..((((((((.((.(((((((((..............))))))).)).)).))))))))..))))))))) ( -36.84) >DroGri_CAF1 121368 96 - 1 ---AUUUGCUGUAUGUGGUCAACAGAGCUGCCAAUUCUGCAGGGCAUGCAGUAGUUGCUUAAAUCAACUGCAAUGCACUACGGAAUUGAAAGUUACAUU ---.....((((.((....))))))((((..((((((((.((.((((...(((((((.......))))))).)))).)).))))))))..))))..... ( -29.00) >DroEre_CAF1 100712 96 - 1 UCCAGA---UGAUUAAGGUCGUCAAAGUUGCCAAAUCCGCAGCGCUUGCAGUAAAUAAGUAAAGACACUGCAAUGCACUACGGAAUUGAAAACUUCAAA ....((---((((....)))))).(((((..(((.((((.((.(((((((((..............))))))).)).)).)))).)))..))))).... ( -29.84) >DroMoj_CAF1 147675 96 - 1 ---AUAUGCUGUCUGUGGUCAACGGAGCUGCCAAUUCUGCAGCGCUUGCAGUAAUUCCUUAAAUCAACUGCAAUGCACUACGGAAUUGAAAGUUACGCA ---...(((..(((((.....)))))(((..((((((((.((.(((((((((..............))))))).)).)).))))))))..)))...))) ( -30.74) >DroAna_CAF1 94286 93 - 1 ---AGU---UGCUCCUGGUCUUCAAAUCUGCCAAGUCCGCAGAGCAUGCAGUUGUAGUUUAAAAACACUGCAAUGCACUACGGAAUUGAAAGUCUCGAA ---...---...((..((..(((((..((....))((((.((.(((((((((.((.........))))))).)))).)).)))).)))))..))..)). ( -22.90) >consensus ___AGA___UGUUUGUGGUCAUCAAAGCUGCCAAUUCCGCAGCGCUUGCAGUAAUUACUUAAAUAAACUGCAAUGCACUACGGAAUUGAAAGUUUCAAA .........................((((..(((.((((.((.(((((((((..............))))))).)).)).)))).)))..))))..... (-20.53 = -20.34 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:25 2006