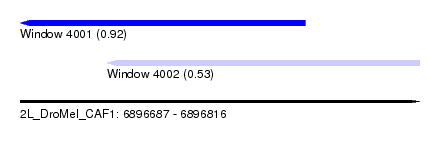
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,896,687 – 6,896,816 |
| Length | 129 |
| Max. P | 0.915500 |

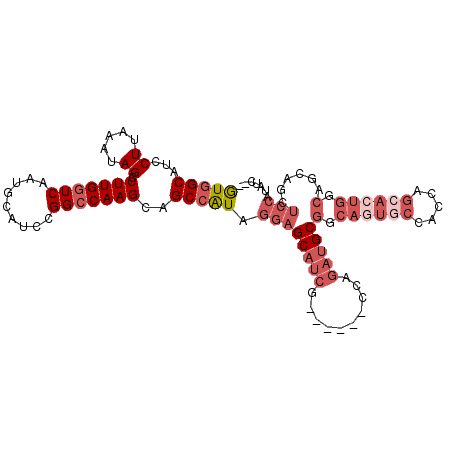
| Location | 6,896,687 – 6,896,779 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915500 |
| Prediction | RNA |
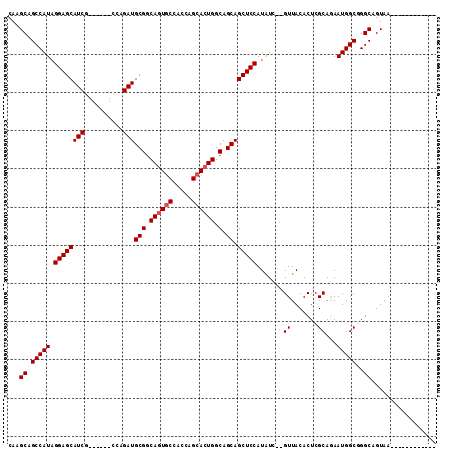
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6896687 92 - 22407834 CAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGCCAGUACCACCAGCACUGGCAGCAGCUCCAUAUC--GUUACACUCGCAGAAUGGCGGGCAGUAA------------ ...((.(((((.((((((((.------...)))((((((((.(.....).)))))).)).))))).....--((.......))...)))))..)).....------------ ( -31.50) >DroSec_CAF1 94683 92 - 1 CAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGGCAGUGCCACCAGCACUGGCAGCAGCUCCAUGUC--GUUACACUCGCAGAAUGGCGGGCAGUAA------------ ...((.(((((.((((((((.------...)))(((.((((((.....)))))).).)).))))).((.(--(.......))))..)))))..)).....------------ ( -35.10) >DroSim_CAF1 88843 92 - 1 CAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGGCAGUGCCACCAGCACUGGCAGCAGCUCCAUAUC--GUUACACUCGCAGAAUGGCGGGCAGUAA------------ ...((.(((((.((((((((.------...)))(((.((((((.....)))))).).)).))))).....--((.......))...)))))..)).....------------ ( -33.50) >DroEre_CAF1 95183 103 - 1 CAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGGCAGUGC-ACCAGCACUGGCAGCAGCUCCAUAUC--GUUACACUCGCAAAAUGGCAGGCAGUAAAAGUGAUGUUUC ...((.(((((.((((((((.------...)))(((.((((((-....)))))).).)).))))).....--((.......))...)))))..))................. ( -32.80) >DroYak_CAF1 93081 107 - 1 CAAGCAGCCAUAGGAGCAUCUCUCCUACCAGAUGCGGCAGUGCCACCAGCAGUGGCAGCAGCUCCAUAUCCAGUUACACUCGCAAAAUGGCAGGCAGUAAAAG-----UUUC ......(((.((((((.....))))))(((..((((((..((((((.....))))))))((((........)))).....))))...)))..)))........-----.... ( -31.20) >consensus CAAGCAGCCAUAGGAGCAUCG______CCAGAUGCGGCAGUGCCACCAGCACUGGCAGCAGCUCCAUAUC__GUUACACUCGCAGAAUGGCGGGCAGUAA____________ ...((.(((((.((((((((..........)))(((.((((((.....)))))).).)).))))).......((.......))...)))))..))................. (-28.70 = -29.10 + 0.40)

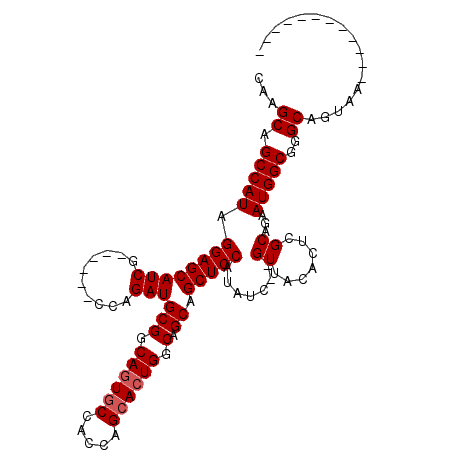
| Location | 6,896,715 – 6,896,816 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -22.58 |
| Energy contribution | -25.13 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6896715 101 - 22407834 GUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGACAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGCCAGUACCACCAGCACUGGCAGCAGCUCCAUAUC-- (((((...........((....))...(((..........)))))))).((((((((.------...)))((((((((.(.....).)))))).)).))))).....-- ( -30.60) >DroSec_CAF1 94711 101 - 1 GUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGCCAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGGCAGUGCCACCAGCACUGGCAGCAGCUCCAUGUC-- (((((...((.....)).(((((((.........)))))))..))))).((((((((.------...)))(((.((((((.....)))))).).)).))))).....-- ( -38.00) >DroSim_CAF1 88871 101 - 1 GUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGCCAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGGCAGUGCCACCAGCACUGGCAGCAGCUCCAUAUC-- (((((...((.....)).(((((((.........)))))))..))))).((((((((.------...)))(((.((((((.....)))))).).)).))))).....-- ( -38.00) >DroEre_CAF1 95223 100 - 1 GUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGCCAAGCAGCCAUAGGAGCAUCG------CCAGAUGCGGCAGUGC-ACCAGCACUGGCAGCAGCUCCAUAUC-- (((((...((.....)).(((((((.........)))))))..))))).((((((((.------...)))(((.((((((-....)))))).).)).))))).....-- ( -38.50) >DroYak_CAF1 93116 109 - 1 GUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGCCAAGCAGCCAUAGGAGCAUCUCUCCUACCAGAUGCGGCAGUGCCACCAGCAGUGGCAGCAGCUCCAUAUCCA (((((...((.....)).(((((((.........)))))))..))))).(((((((((........))))))(((.(((((((.....)))))).).))).....))). ( -36.20) >DroAna_CAF1 89687 80 - 1 AUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGCCAAGCAGCAGUGGAAGCAAAG------GCAAAGGCGACUGUGUCUAC---------------------UU-- ..(((((.........(((((.(((..(((.(((.((......))...))).)))..)------)).)))))....)))))...---------------------..-- ( -21.92) >consensus GUGGCAUCCUUAAAUAGCCUUGGUCAAUGCAUCCGGCCAAGCAGCCAUAGGAGCAUCG______CCAGAUGCGGCAGUGCCACCAGCACUGGCAGCAGCUCCAUAUC__ (((((...((.....)).(((((((.........)))))))..))))).((((((((..........)))))(.((((((.....)))))).)......)))....... (-22.58 = -25.13 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:21 2006