
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,862,377 – 6,862,597 |
| Length | 220 |
| Max. P | 0.938434 |

| Location | 6,862,377 – 6,862,488 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -24.83 |
| Energy contribution | -25.13 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6862377 111 - 22407834 CCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUAAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGGUGAGUUU-- (((((...........(((((((((......)))))))))........(((.((((...(((((........)))))......)))).)))........))))).......-- ( -40.00) >DroVir_CAF1 97088 112 - 1 CCUGGUGGUGCUGACCGUGGCCGCCAAUUUCGGUGGCUAUCUGAUAUCGUUGACCACCCUGCCACGUCUGAUUGGUAUGCUGCUGGUGGGCAUACUCUUCCAGGUGAGUC-GC (((((..(((((.((((..((.(((((((...(((((....((...........))....)))))....)))))))..))..).))).)))))......)))))......-.. ( -40.70) >DroPse_CAF1 84174 101 - 1 CCUGGUCGUCCUAACUGUGGCAGCCAACUUUGGCGGCUACCUGAUAUCUCUGACCACCCUGCCUCGUCUGAUUGGCAUGCUCCUGGUGGGCAUACUCUUUC------------ ..((((((........(((((.((((....)))).)))))..........))))))....(((..........)))((((.((....))))))........------------ ( -28.67) >DroWil_CAF1 53224 109 - 1 UCUUGUUGUGCUGACUGUGGCAGCCAAUUUCGGUGGCUAUUUGAUAUCAUUGACCACAUUACCAAGAUUAAUUGGUAUGCUCAUGGUGGGCAUAUUAUUUCAGGUAAGA---- .((((.((((((.(((((((((..........(((((....((....))..).))))..((((((......))))))))).)))))).))))))......)))).....---- ( -31.40) >DroMoj_CAF1 98593 111 - 1 CCUGGUGGUGCUCACGGUGGCCGCCAAUUUCGGUGGCUACAUAAUAUCACUGACCACUCUGCCUCGUCUCAUUGGCAUGCUGCUGGUGGGCAUACUCUUUCAGGUGAGA--AC ..((((((((......(((((((((......))))))))).......)))).))))(((.((((.(((.(((..((.....))..))))))..........))))))).--.. ( -39.82) >DroAna_CAF1 61441 111 - 1 UCUCGUCGUCCUUACUGUGGCCGCCAACUUUGGCGGCUACGUGAUAUCGCUGACUACCCUUCCCCGACUAAUUGGCAUGCUCCUUGUGGGCAUCCUAUUCCAGGUGAGUGU-- .(((..((((......((((((((((....))))))))))(((....)))...............))).....((.((((.((....)))))))).......)..)))...-- ( -37.90) >consensus CCUGGUCGUGCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUGAUAUCGCUGACCACCCUGCCACGUCUAAUUGGCAUGCUCCUGGUGGGCAUACUCUUCCAGGUGAGU____ (((((...........(((((((((......)))))))))..........((.(((((.((((..........)))).......))))).)).......)))))......... (-24.83 = -25.13 + 0.31)

| Location | 6,862,408 – 6,862,528 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -27.51 |
| Energy contribution | -26.57 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6862408 120 - 22407834 UCAUCGGCGAUUCGGCAGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUAAUAUCGCUGACCACCUUGCCACGUCUCAUUGGCAUGC ...((((((((.((((.((((.....)))).(((....)))...))))........(((((((((......))))))))).....))))))))......(((((........)))))... ( -44.80) >DroVir_CAF1 97120 120 - 1 UUAUCGGGGACACAGCUGCGCCGGGCGGUCAACUGUUUAGCCUGGUGGUGCUGACCGUGGCCGCCAAUUUCGGUGGCUAUCUGAUAUCGUUGACCACCCUGCCACGUCUGAUUGGUAUGC .(((((....).((((((.((.(((.(((((((.(((..(((....)))...))).(((((((((......)))))))))........))))))).))).)))).).)))...))))... ( -47.70) >DroGri_CAF1 85169 120 - 1 UUAUUGGGGACACCGCUGCCCCUGGCGGGCAGCUCUUCAGUCUAGUGGUGCUUACAGUAGCUGCCAAUUUUGGUGGUUAUCUGAUAUCAUUGACCACCCUGCCAAGGUUGAUUGGUAUGC .....((((.....(((((((.....)))))))......(((.(((((((....(((((((..(((....)))..)))).))).))))))))))..))))(((((......))))).... ( -44.80) >DroWil_CAF1 53253 120 - 1 UCAUUGGAGACUCAGCUGCCCCUGGUGGACAGCUUUUUGGUCUUGUUGUGCUGACUGUGGCAGCCAAUUUCGGUGGCUAUUUGAUAUCAUUGACCACAUUACCAAGAUUAAUUGGUAUGC ......((((((.(((((((......)).)))))....))))))((((.((((.(....)))))))))....(((((....((....))..).))))..((((((......))))))... ( -32.10) >DroMoj_CAF1 98624 120 - 1 UCAUUGGGGACACAGCUGCCCCGGGUGGGCAGCUCUUCAGCCUGGUGGUGCUCACGGUGGCCGCCAAUUUCGGUGGCUACAUAAUAUCACUGACCACUCUGCCUCGUCUCAUUGGCAUGC (((...(((((..((((((((.....))))))))...(((..((((((((......(((((((((......))))))))).......)))).))))..)))....)))))..)))..... ( -50.22) >DroAna_CAF1 61472 120 - 1 UCAUUGGCGACUCGGCUGCCCCUGGCGGACAGCUCUUCGGUCUCGUCGUCCUUACUGUGGCCGCCAACUUUGGCGGCUACGUGAUAUCGCUGACUACCCUUCCCCGACUAAUUGGCAUGC ....((.(((.(((((((((...))))).((((.....((.(.....).))((((.((((((((((....))))))))))))))....))))...........))))....))).))... ( -41.10) >consensus UCAUUGGGGACACAGCUGCCCCUGGCGGGCAGCUCUUCAGCCUGGUGGUGCUCACUGUGGCCGCCAAUUUCGGUGGCUACCUGAUAUCACUGACCACCCUGCCACGUCUAAUUGGCAUGC .....((......(((((.(((....))))))))..(((((......((....)).(((((((((......)))))))))........)))))))....(((((........)))))... (-27.51 = -26.57 + -0.94)


| Location | 6,862,448 – 6,862,568 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
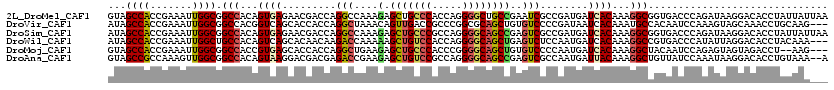
>2L_DroMel_CAF1 6862448 120 + 22407834 GUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCACCAGGGGCUGCCGAAUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACACCUAUUAUUAA ...(((...((.((((((((((...((....)).....))))...(.((.((((.....)))).))).....))))))..)).....)))((((.((.......)).))))......... ( -37.80) >DroVir_CAF1 97160 117 + 1 AUAGCCACCGAAAUUGGCGGCCACGGUCAGCACCACCAGGCUAAACAGUUGACCGCCCGGCGCAGCUGUGUCCCCGAUAAUCACAAAUGCCACAAUCCAAAGUAGCAAACCUGCAAG--- ...((((.......)))).(((.((((((((................))))))))...)))((((.((((...........))))..(((.((........)).)))...))))...--- ( -31.19) >DroSim_CAF1 59405 120 + 1 AUAGCCACCGAAAUUGGCGGCCACAGUGAGAACGACCAGGCCAAAGAGCUGCCCGCCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACACCUAUUAUUAA ...(((...((.((((((((((...((....)).....))))...(.(((((((.....)))))))).....))))))..)).....)))((((.((.......)).))))......... ( -42.60) >DroWil_CAF1 53293 117 + 1 AUAGCCACCGAAAUUGGCUGCCACAGUCAGCACAACAAGACCAAAAAGCUGUCCACCAGGGGCAGCUGAGUCUCCAAUGAUCACAAAGGCCGUGACCCAUAUUAGGACACCUACAAA--- .((((((.......)))))).....(((.........((((.....((((((((.....))))))))..))))...(((.((((.......))))..))).....))).........--- ( -29.70) >DroMoj_CAF1 98664 115 + 1 GUAGCCACCGAAAUUGGCGGCCACCGUGAGCACCACCAGGCUGAAGAGCUGCCCACCCGGGGCAGCUGUGUCCCCAAUGAUCACAAAGGCUACAAUCCAGAGUAGUAGACCU--AAG--- ((((((...((.((((((((((...(((.....)))..))))).(.((((((((.....)))))))).)....)))))..)).....))))))...................--...--- ( -39.90) >DroAna_CAF1 61512 118 + 1 GUAGCCGCCAAAGUUGGCGGCCACAGUAAGGACGACGAGACCGAAGAGCUGUCCGCCAGGGGCAGCCGAGUCGCCAAUGAUUACAAAGGCUGUUAUCCAAAUAAGGACACCUGUAAA--A ((.(((((((....))))))).)).....(((..(((.(((((....(((((((.....))))))))).)))(((..((....))..))))))..))).....(((...))).....--. ( -42.40) >consensus AUAGCCACCGAAAUUGGCGGCCACAGUGAGCACGACCAGGCCAAAGAGCUGCCCACCAGGGGCAGCCGAGUCGCCAAUGAUCACAAAGGCCGUGACCCAGAUAAGGACACCUAUAAA___ ...((((.......)))).(((...((((.........(((....(.(((((((.....))))))))..)))........))))...))).............................. (-22.16 = -22.38 + 0.22)
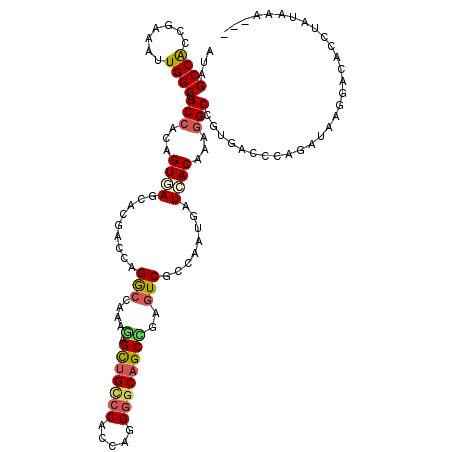
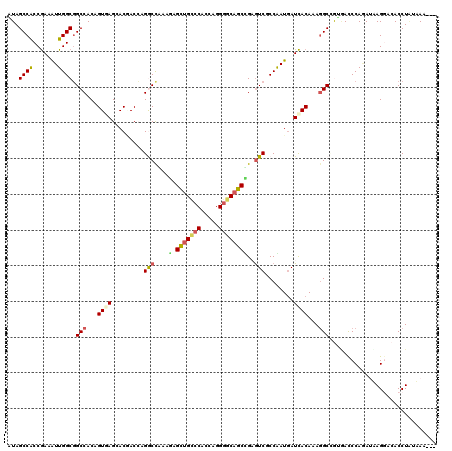
| Location | 6,862,448 – 6,862,568 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.64 |
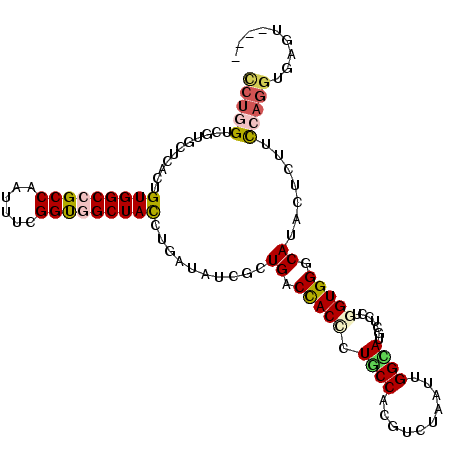
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6862448 120 - 22407834 UUAAUAAUAGGUGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGAUUCGGCAGCCCCUGGUGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUAC .........((((.......(..(((((.((((..((....))..))))....(((.((((.....)))).)))...)))))..).......))))(((((((((......))))))))) ( -46.94) >DroVir_CAF1 97160 117 - 1 ---CUUGCAGGUUUGCUACUUUGGAUUGUGGCAUUUGUGAUUAUCGGGGACACAGCUGCGCCGGGCGGUCAACUGUUUAGCCUGGUGGUGCUGACCGUGGCCGCCAAUUUCGGUGGCUAU ---.(..((((..((((((........))))))))))..).....(....).((((..((((((((.............))))))))..))))...(((((((((......))))))))) ( -47.62) >DroSim_CAF1 59405 120 - 1 UUAAUAAUAGGUGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGCGGGCAGCUCUUUGGCCUGGUCGUUCUCACUGUGGCCGCCAAUUUCGGUGGCUAU .........((((.......(..(((((.((((..((....))..))))....((((((((.....))))))))...)))))..).......))))(((((((((......))))))))) ( -51.24) >DroWil_CAF1 53293 117 - 1 ---UUUGUAGGUGUCCUAAUAUGGGUCACGGCCUUUGUGAUCAUUGGAGACUCAGCUGCCCCUGGUGGACAGCUUUUUGGUCUUGUUGUGCUGACUGUGGCAGCCAAUUUCGGUGGCUAU ---.......(((.(((.....))).))).(((...((.(.(((..((((((.(((((((......)).)))))....))))))...))).).))...)))(((((.......))))).. ( -36.00) >DroMoj_CAF1 98664 115 - 1 ---CUU--AGGUCUACUACUCUGGAUUGUAGCCUUUGUGAUCAUUGGGGACACAGCUGCCCCGGGUGGGCAGCUCUUCAGCCUGGUGGUGCUCACGGUGGCCGCCAAUUUCGGUGGCUAC ---...--(((.((((...........))))))).(((((((((..((.....((((((((.....))))))))......))..))))...)))))(((((((((......))))))))) ( -48.40) >DroAna_CAF1 61512 118 - 1 U--UUUACAGGUGUCCUUAUUUGGAUAACAGCCUUUGUAAUCAUUGGCGACUCGGCUGCCCCUGGCGGACAGCUCUUCGGUCUCGUCGUCCUUACUGUGGCCGCCAACUUUGGCGGCUAC .--....(((((((((......))))).(((((.((((........))))...)))))..))))..((((.((.(....)....)).)))).....((((((((((....)))))))))) ( -44.70) >consensus ___AUUAUAGGUGUCCUAAUCUGGAUCACAGCCUUUGUGAUCAUCGGCGACUCAGCUGCCCCUGGCGGGCAGCUCUUUGGCCUGGUCGUGCUCACUGUGGCCGCCAAUUUCGGUGGCUAC .........((((.......((((((((...((..((....))..))......(((((.(((....))))))))...)))))))).......))))(((((((((......))))))))) (-29.30 = -29.72 + 0.42)
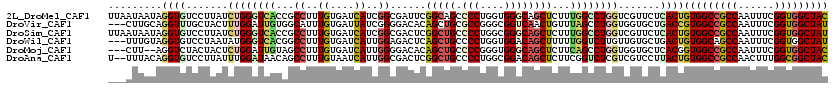
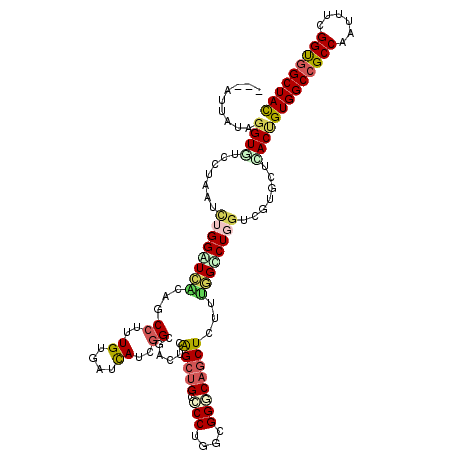

| Location | 6,862,488 – 6,862,597 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6862488 109 + 22407834 CCAAAGAGCUGCCCACCAGGGGCUGCCGAAUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACACCUAUUAUUAAGGAA---AUAGUUAACACAAUUAU---AAUGUAUG ((...(.((.((((.....)))).))).....(((..((....))..)))((((.((.......)).)))).........))..---((((((.....))))))---........ ( -24.00) >DroSec_CAF1 66818 106 + 1 CCAAAGAGCUGCCCACCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACUCCUAUUAUUAAGAAA---AUAGUUAAUACAAUUAU---AA---CUU .....(.(((((((.....))))))))((((((((..((....))..)))(....).........)))))(((((........)---)))).............---..---... ( -29.50) >DroEre_CAF1 66630 108 + 1 CCAAAGAGCUGCCCUCCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUGAGGACACCUGUGAUUUAGGAA---AUAAUU-ACACAAUUAU---AACCCCAU ((...(.(((((((.....))))))))((((((((..((....))..)))((((.(((....).)).))))...))))).))..---((((((-....))))))---........ ( -33.30) >DroWil_CAF1 53333 97 + 1 CCAAAAAGCUGUCCACCAGGGGCAGCUGAGUCUCCAAUGAUCACAAAGGCCGUGACCCAUAUUAGGACACCUACAAA------C---AAAGUUUUGCCAAUUAU---UA------ .(((((((((((((.....))))))))..((((...(((.((((.......))))..)))....)))).........------.---....)))))........---..------ ( -22.20) >DroYak_CAF1 64647 107 + 1 CCAAAGAGCUGCCCUCCAGGGGCAGCCGAGUCGCCGAUGAUCACGAAGGCGGUGACCCAGAUAAGGACACCUGUAAUUCAGAA----AAAUUU-AUAUAUUUAU---AACCUCAA .....(.(((((((.....))))))))(.(((((((.(........)..))))))).).....(((....(((.....)))..----....((-(((....)))---)))))... ( -29.80) >DroAna_CAF1 61552 113 + 1 CCGAAGAGCUGUCCGCCAGGGGCAGCCGAGUCGCCAAUGAUUACAAAGGCUGUUAUCCAAAUAAGGACACCUGUAAA--AGGGUAUUAUAAUAAAUUAAAUUAGAGAAAACUAAA ..((.(.(((((((.....))))))))...))(((..((....))..)))((((((((......))).((((.....--.))))....))))).......((((......)))). ( -24.40) >consensus CCAAAGAGCUGCCCACCAGGGGCAGCCGAGUCGCCGAUGAUCACAAAGGCGGUGACCCAGAUAAGGACACCUAUAAUU_AGGAA___AUAGUUAAUACAAUUAU___AA_CUCAA .....(.(((((((.....)))))))).....(((..((....))..)))((((.((.......)).))))............................................ (-19.91 = -20.13 + 0.23)

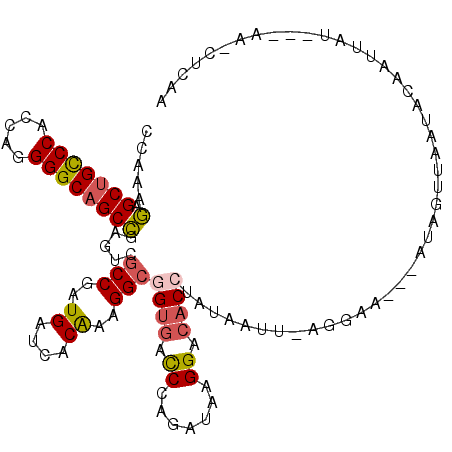
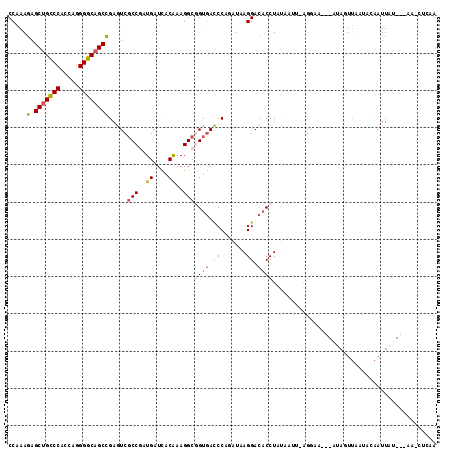
| Location | 6,862,488 – 6,862,597 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -21.37 |
| Energy contribution | -22.90 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6862488 109 - 22407834 CAUACAUU---AUAAUUGUGUUAACUAU---UUCCUUAAUAAUAGGUGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGAUUCGGCAGCCCCUGGUGGGCAGCUCUUUGG .(((((..---.....))))).......---.............((((.(((.....))).))))(((..((....))..))).....(((.((((.....)))).)))...... ( -27.80) >DroSec_CAF1 66818 106 - 1 AAG---UU---AUAAUUGUAUUAACUAU---UUUCUUAAUAAUAGGAGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGUGGGCAGCUCUUUGG ...---..---.............((((---(........)))))(((((.......((....))(((..((....))..))))))))((((((((.....))))))))...... ( -32.00) >DroEre_CAF1 66630 108 - 1 AUGGGGUU---AUAAUUGUGU-AAUUAU---UUCCUAAAUCACAGGUGUCCUCAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGAGGGCAGCUCUUUGG .(((((..---((((((....-))))))---.)))))..((...((((.(((.....))).))))(((..((....))..)))))...((((((((.....))))))))...... ( -36.50) >DroWil_CAF1 53333 97 - 1 ------UA---AUAAUUGGCAAAACUUU---G------UUUGUAGGUGUCCUAAUAUGGGUCACGGCCUUUGUGAUCAUUGGAGACUCAGCUGCCCCUGGUGGACAGCUUUUUGG ------..---.......(((((.....---.------))))).(((.(((.......(((((((.....)))))))...))).))).(((((((......)).)))))...... ( -22.80) >DroYak_CAF1 64647 107 - 1 UUGAGGUU---AUAAAUAUAU-AAAUUU----UUCUGAAUUACAGGUGUCCUUAUCUGGGUCACCGCCUUCGUGAUCAUCGGCGACUCGGCUGCCCCUGGAGGGCAGCUCUUUGG ..(((...---..........-......----............((((.(((.....))).))))(((............)))..)))((((((((.....))))))))...... ( -32.60) >DroAna_CAF1 61552 113 - 1 UUUAGUUUUCUCUAAUUUAAUUUAUUAUAAUACCCU--UUUACAGGUGUCCUUAUUUGGAUAACAGCCUUUGUAAUCAUUGGCGACUCGGCUGCCCCUGGCGGACAGCUCUUCGG .((((......))))..................(((--.....)))(((((......)))))...(((..((....))..)))((...(((((((......)).)))))..)).. ( -22.00) >consensus AUGAG_UU___AUAAUUGUAUUAACUAU___UUCCU_AAUUACAGGUGUCCUUAUCUGGGUCACCGCCUUUGUGAUCAUCGGCGACUCGGCUGCCCCUGGUGGGCAGCUCUUUGG ............................................((((.(((.....))).))))(((..((....))..))).....((((((((.....))))))))...... (-21.37 = -22.90 + 1.53)
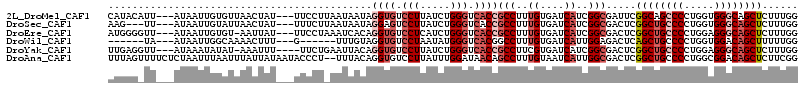
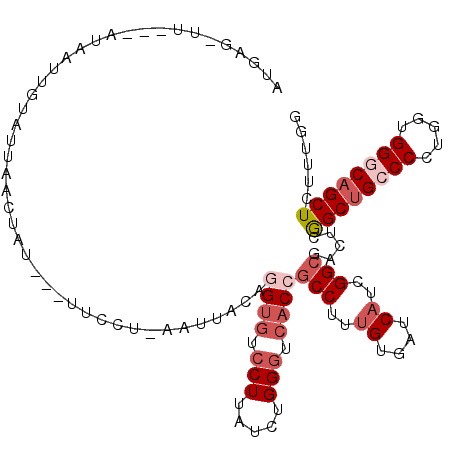
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:13 2006