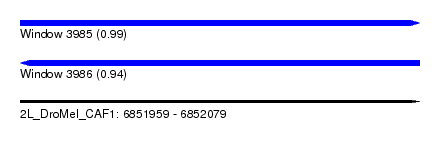
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,851,959 – 6,852,079 |
| Length | 120 |
| Max. P | 0.991167 |

| Location | 6,851,959 – 6,852,079 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -21.32 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
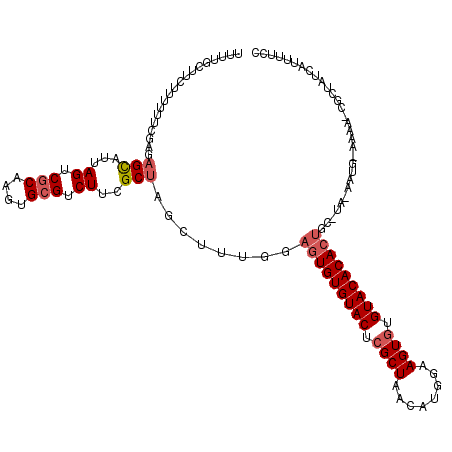
>2L_DroMel_CAF1 6851959 120 + 22407834 UUUUACUUCUUUUUUCGAGAGCAUUAGUCGCAAGUGCGUCUUCGCUAGCUUUGGAAUGUGUACUCGCUAAUAUGGAAGUAUGUACACACUGCAUAGAAUGAAAAAACGCUAUCUUAUUCC .........((((((((..(((...((.(((....))).))..)))..((.((.(.(((((((..(((........)))..))))))).).)).))..)))))))).............. ( -26.70) >DroSec_CAF1 56332 104 + 1 UUUUGCUUCUUUUUUCGAGAGCAUUAGUCGCAAGUGCGUCUUCGCUAACUUUGGAGUGUGUACUCGCUAACAUGAAAGUGUGUACACACU----------------CGCUAUCAUUUUCC ...((((.(((.....)))))))......(((((((((....)))..))))..((((((((((.((((........)))).)))))))))----------------)))........... ( -31.90) >DroSim_CAF1 48083 104 + 1 UUUUGUUUCUUUUUUCGAGCGCAUUAGUCGCAAGUGCGUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAAAAUGGAAGUGUGUACACACU----------------CGCUAUCAUUUUCC .................(((((...((.(((....))).))..))........((((((((((.((((........)))).)))))))))----------------)))).......... ( -32.60) >DroEre_CAF1 55968 118 + 1 UUUUGCCACUUUUU--CAGAGCUCAAGUCGCAAGUGCGUCUUUGCUAGCAUUGGAGUGUGUACUCGCUAACUCGAAAGUGUGUACACACUGCCUAGAAUGUAAAAUCUUGUACAUCCUUC ..((((.((((...--........)))).))))(((((..(((((...(...(((((((((((.((((........)))).))))))))).))..)...)))))....)))))....... ( -32.80) >DroYak_CAF1 54093 118 + 1 UUUUGCUCCUUUUU--GAGAGUUUGAGUCUCGAAUGCGUCUUUGCUAACGUUGGAGUGUGUACUCGCUAACUGGGCAGUGUGUACACACCGCAUACAAUGUAAAAUCACUUACAUUUUCC ............((--((((.......))))))((((((((..((....)).)))((((((((.((((........)))).)))))))))))))..(((((((......))))))).... ( -32.60) >consensus UUUUGCUUCUUUUUUCGAGAGCAUUAGUCGCAAGUGCGUCUUCGCUAGCUUUGGAGUGUGUACUCGCUAACAUGGAAGUGUGUACACACUGC_UA_AAUG_AAAA_CGCUAUCAUUUUCC ...................(((...((.(((....))).))..)))........(((((((((.((((........)))).))))))))).............................. (-21.32 = -22.16 + 0.84)

| Location | 6,851,959 – 6,852,079 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.58 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
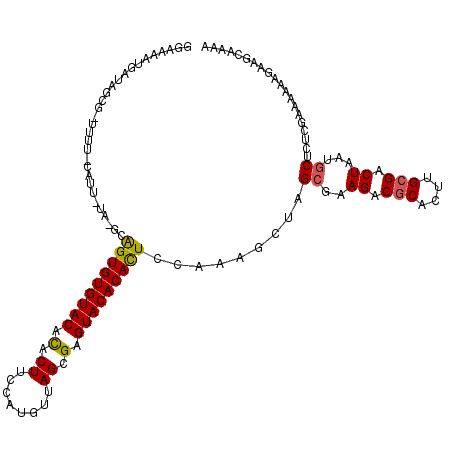
>2L_DroMel_CAF1 6851959 120 - 22407834 GGAAUAAGAUAGCGUUUUUUCAUUCUAUGCAGUGUGUACAUACUUCCAUAUUAGCGAGUACACAUUCCAAAGCUAGCGAAGACGCACUUGCGACUAAUGCUCUCGAAAAAAGAAGUAAAA .......((.((((((...((......((.(((((((((...((........))...))))))))).))..((..(((....)))....))))..)))))).))................ ( -23.90) >DroSec_CAF1 56332 104 - 1 GGAAAAUGAUAGCG----------------AGUGUGUACACACUUUCAUGUUAGCGAGUACACACUCCAAAGUUAGCGAAGACGCACUUGCGACUAAUGCUCUCGAAAAAAGAAGCAAAA ......(((.((((----------------(((((((((.(.((........)).).))))))))))....((((((((((.....))).)).)))))))).)))............... ( -27.80) >DroSim_CAF1 48083 104 - 1 GGAAAAUGAUAGCG----------------AGUGUGUACACACUUCCAUUUUAGCGAGUACACACUCCAAAGCUAGCGAAGACGCACUUGCGACUAAUGCGCUCGAAAAAAGAAACAAAA .........(((((----------------(((((((((.(.((........)).).))))))))))....))))(((.((.(((....))).))....))).................. ( -27.50) >DroEre_CAF1 55968 118 - 1 GAAGGAUGUACAAGAUUUUACAUUCUAGGCAGUGUGUACACACUUUCGAGUUAGCGAGUACACACUCCAAUGCUAGCAAAGACGCACUUGCGACUUGAGCUCUG--AAAAAGUGGCAAAA ..((((((((........)))))))).((.(((((((((.(.((........)).).)))))))))))..(((((((.(((.(((....))).)))..)).((.--....)))))))... ( -35.10) >DroYak_CAF1 54093 118 - 1 GGAAAAUGUAAGUGAUUUUACAUUGUAUGCGGUGUGUACACACUGCCCAGUUAGCGAGUACACACUCCAACGUUAGCAAAGACGCAUUCGAGACUCAAACUCUC--AAAAAGGAGCAAAA ....((((((((....))))))))...(((.((((((((.(.(((......))).).))))))))(((..((((......)))).....((((.......))))--.....))))))... ( -30.40) >consensus GGAAAAUGAUAGCG_UUUU_CAUU_UA_GCAGUGUGUACACACUUCCAUGUUAGCGAGUACACACUCCAAAGCUAGCGAAGACGCACUUGCGACUAAUGCUCUCGAAAAAAGAAGCAAAA ..............................(((((((((.(.((........)).).))))))))).........((..((.(((....))).))...)).................... (-15.66 = -15.58 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:06 2006