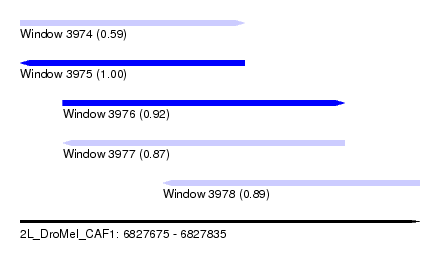
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,827,675 – 6,827,835 |
| Length | 160 |
| Max. P | 0.999852 |

| Location | 6,827,675 – 6,827,765 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -12.22 |
| Consensus MFE | -1.88 |
| Energy contribution | -1.32 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.15 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
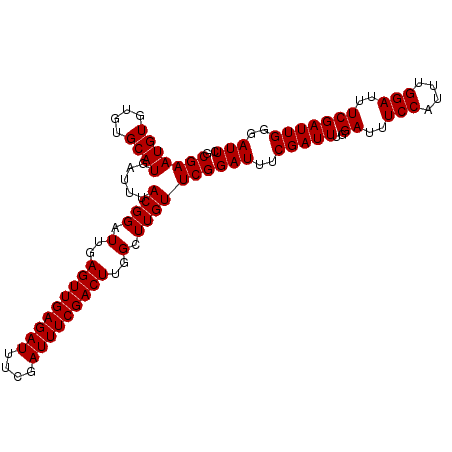
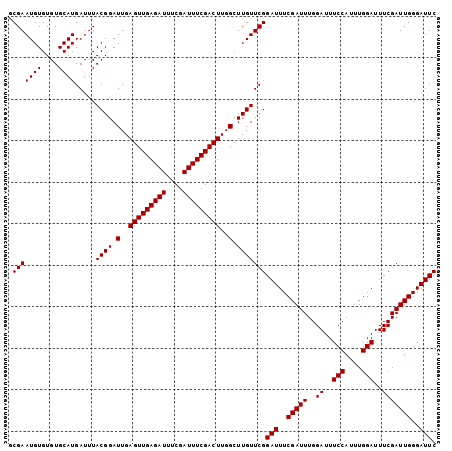
>2L_DroMel_CAF1 6827675 90 + 22407834 GGGCGAUCCAAAUCCAAAUCGGAAACCCAAUCGGAAUCCAAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAA (((((((.....(((.....))).........(((.(((.................))).)))..))))...(((....)))..)))... ( -14.63) >DroPse_CAF1 40880 75 + 1 GGGGAAUGUGAAUGGAAAUGG--------AUUGAAAU-------UAAAUCCAAAUCGAAAUCCCAACUGAAAUCCAAAUCGAAAUUGAAA .((((...(((((....))((--------(((.....-------..)))))...)))...)))).......................... ( -12.00) >DroSec_CAF1 31826 90 + 1 GCGCAAUCCAAAUCCAAAUCGGAAACCCAAUCGGAAUCCCAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAA ......((((..(((.....))).........(((.(((.................))).)))...............))))........ ( -11.23) >DroSim_CAF1 23684 90 + 1 GCGCAAUCCAAAUCCAAAUCGGAAACCCAAUCGGAAUCCCAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAA ......((((..(((.....))).........(((.(((.................))).)))...............))))........ ( -11.23) >DroPer_CAF1 32734 75 + 1 GGGGAAUGUGAAUGGAAAUGG--------AUUGAAAU-------UAAAUCCAAAUCGAAAUCCCAACUGAAAUCCAAAUCGAAAUUGAAA .((((...(((((....))((--------(((.....-------..)))))...)))...)))).......................... ( -12.00) >consensus GGGCAAUCCAAAUCCAAAUCGGAAACCCAAUCGGAAUCC_AAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAA ......................................................((((........))))........(((....))).. ( -1.88 = -1.32 + -0.56)

| Location | 6,827,675 – 6,827,765 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -16.54 |
| Energy contribution | -20.70 |
| Covariance contribution | 4.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.20 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6827675 90 - 22407834 UUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUUGGAUUCCGAUUGGGUUUCCGAUUUGGAUUUGGAUCGCCC ...(((.(((((.(((((.(((.....))).))))).))))).)))((((..(((((.((((((....)))))).)))))..)))).... ( -34.10) >DroPse_CAF1 40880 75 - 1 UUUCAAUUUCGAUUUGGAUUUCAGUUGGGAUUUCGAUUUGGAUUUA-------AUUUCAAU--------CCAUUUCCAUUCACAUUCCCC .(((((.......)))))........((((....((..((((((..-------.....)))--------)))..)).........)))). ( -11.62) >DroSec_CAF1 31826 90 - 1 UUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUGGGAUUCCGAUUGGGUUUCCGAUUUGGAUUUGGAUUGCGC ((..((..((((.(((((.(((((((((((((((((((((....)))))))))))))))))))))..)))))..))))))..))...... ( -36.80) >DroSim_CAF1 23684 90 - 1 UUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUGGGAUUCCGAUUGGGUUUCCGAUUUGGAUUUGGAUUGCGC ((..((..((((.(((((.(((((((((((((((((((((....)))))))))))))))))))))..)))))..))))))..))...... ( -36.80) >DroPer_CAF1 32734 75 - 1 UUUCAAUUUCGAUUUGGAUUUCAGUUGGGAUUUCGAUUUGGAUUUA-------AUUUCAAU--------CCAUUUCCAUUCACAUUCCCC .(((((.......)))))........((((....((..((((((..-------.....)))--------)))..)).........)))). ( -11.62) >consensus UUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUU_GGAUUCCGAUUGGGUUUCCGAUUUGGAUUUGGAUUGCCC ...........((((((((((.((((((((.((((((((((((((......)))))))))))))).)))))))).))))))))))..... (-16.54 = -20.70 + 4.16)

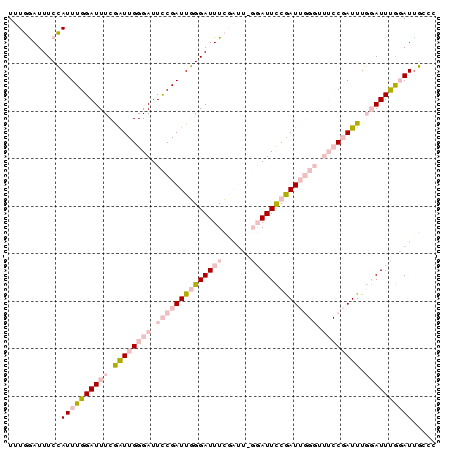
| Location | 6,827,692 – 6,827,805 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 98.82 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6827692 113 + 22407834 AUCGGAAACCCAAUCGGAAUCCAAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAAUCGAAAUCCGAACAACCCAAGUCGAAAUCGAAAUCUCAAC .(((((...((....))........((((..(((.....))).........((..(((....)))..))....))))..)))))..........(((....)))......... ( -17.50) >DroSec_CAF1 31843 113 + 1 AUCGGAAACCCAAUCGGAAUCCCAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAAUCGAAAUCCGAACAAGCCAAGUCGAAAUCGAAAUCUCAAC .(((((.........((....))..((((..(((.....))).........((..(((....)))..))....))))..)))))..........(((....)))......... ( -18.10) >DroSim_CAF1 23701 113 + 1 AUCGGAAACCCAAUCGGAAUCCCAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAAUCGAAAUCCGAACAAGCCAAGUCGAAAUCGAAAUCUCAAC .(((((.........((....))..((((..(((.....))).........((..(((....)))..))....))))..)))))..........(((....)))......... ( -18.10) >consensus AUCGGAAACCCAAUCGGAAUCCCAAUCGAAAUCCCAAUCGGAAUCCCAAUCGAAAUCCAAAUGGAAAUCCAAAUCGAAAUCCGAACAAGCCAAGUCGAAAUCGAAAUCUCAAC .(((((...((....))........((((..(((.....))).........((..(((....)))..))....))))..)))))..........(((....)))......... (-17.50 = -17.50 + 0.00)

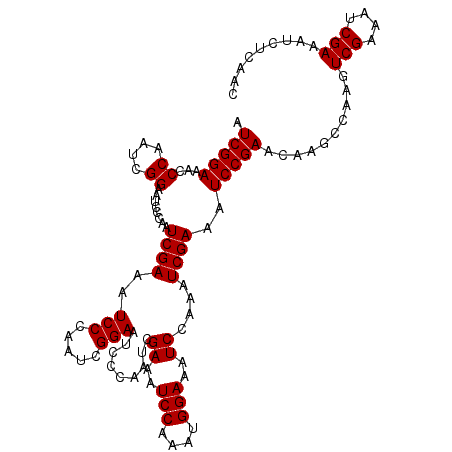
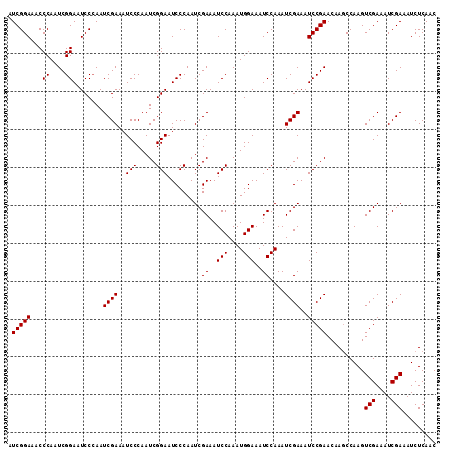
| Location | 6,827,692 – 6,827,805 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 98.82 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -43.03 |
| Energy contribution | -43.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -6.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6827692 113 - 22407834 GUUGAGAUUUCGAUUUCGACUUGGGUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUUGGAUUCCGAUUGGGUUUCCGAU (((((((((.(((...((((....)))).))))))))))))(((((..((((.(((((.((((((((..(((((....)))))..))).))))).))))).))))..))))). ( -44.70) >DroSec_CAF1 31843 113 - 1 GUUGAGAUUUCGAUUUCGACUUGGCUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUGGGAUUCCGAUUGGGUUUCCGAU (((((((((.(((...(((......))).))))))))))))(((((..((((.((((((..((((((..(((((....)))))..))))))..).))))).))))..))))). ( -44.10) >DroSim_CAF1 23701 113 - 1 GUUGAGAUUUCGAUUUCGACUUGGCUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUGGGAUUCCGAUUGGGUUUCCGAU (((((((((.(((...(((......))).))))))))))))(((((..((((.((((((..((((((..(((((....)))))..))))))..).))))).))))..))))). ( -44.10) >consensus GUUGAGAUUUCGAUUUCGACUUGGCUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUCCGAUUGGGAUUUCGAUUGGGAUUCCGAUUGGGUUUCCGAU (((((((((.(((...(((......))).))))))))))))(((((..((((.((((((((((((((..(((((....)))))..))))))))).))))).))))..))))). (-43.03 = -43.37 + 0.33)


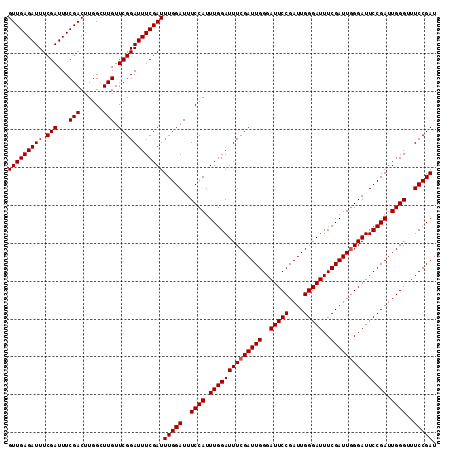
| Location | 6,827,732 – 6,827,835 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 98.71 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6827732 103 - 22407834 GCGAAUGUGUGUGCAUGAUUUACGGAUUGAGUUGAGAUUUCGAUUUCGACUUGGGUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUC .(((((((....)))).....((((.(..(((((((((....)))))))))..).)))))))(((..(((((..((..(((....)))..)))))))..))). ( -27.30) >DroSec_CAF1 31883 103 - 1 ACGAAUGUGUGUGCAUGAUUUACGGAUUGAGUUGAGAUUUCGAUUUCGACUUGGCUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUC .(((((((....)))).....(((..(..(((((((((....)))))))))..)..))))))(((..(((((..((..(((....)))..)))))))..))). ( -27.70) >DroSim_CAF1 23741 103 - 1 GCGAAUGUGUGUGCAUGAUUUACGGAUUGAGUUGAGAUUUCGAUUUCGACUUGGCUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUC .(((((((....)))).....(((..(..(((((((((....)))))))))..)..))))))(((..(((((..((..(((....)))..)))))))..))). ( -27.40) >consensus GCGAAUGUGUGUGCAUGAUUUACGGAUUGAGUUGAGAUUUCGAUUUCGACUUGGCUUGUUCGGAUUUCGAUUUGGAUUUCCAUUUGGAUUUCGAUUGGGAUUC .(((((((....)))).....((((.(..(((((((((....)))))))))..).)))))))(((..(((((..((..(((....)))..)))))))..))). (-27.40 = -27.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:58 2006