
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,788,359 – 6,788,471 |
| Length | 112 |
| Max. P | 0.668614 |

| Location | 6,788,359 – 6,788,471 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.57 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -15.69 |
| Energy contribution | -17.17 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
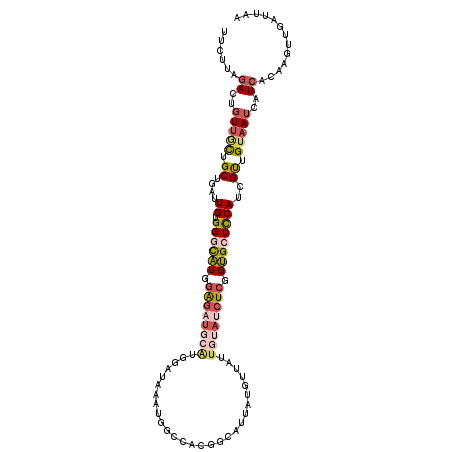
>2L_DroMel_CAF1 6788359 112 + 22407834 UUCUUAGACUGUUGCUGCUGAUUGUGGGCAUGGAGAUGCACGGAUAAAUGGCCACGGCAUUAUGUUAUUGUAUCUCGGUGCUCCAUCGUUGUAAUCAUCACAAGUUGAUUAA ......((..(((((.((.(((.(.((((((.((((((((..((((....((....))....))))..)))))))).)))))))))))).)))))..))............. ( -34.40) >DroSec_CAF1 55215 91 + 1 UUCUUAGACUGUUGCUGCUGAUUGUGGGCAUGGAG---------------------GCAUUAUGUUAUUGUAUCUCGGUGCUCCAUCGUUGUAAUCAUCACAAGUUUAUUAA ....(((((((((((.((.(((.(.((((((.(((---------------------..((.........))..))).)))))))))))).))))).......)))))).... ( -21.71) >DroSim_CAF1 57032 112 + 1 UUCUUGGACUGUUGCUGCUGAUUGUGGGCAUGGAGAUGCACGGAUAAAUGGCCACGGCAUUAUAUUAUUGUAUCUCGGUGCUCCAUCGUUGUAAUCAUCACAAGUUGAUUAA ..((((((..(((((.((.(((.(.((((((.((((((((..((((....((....))....))))..)))))))).)))))))))))).)))))..)).))))........ ( -37.80) >DroEre_CAF1 57865 112 + 1 UUCUUGGACUGUUGCUGCUGAUUGUGGGCAUGGAGAUGCAUGGAUAAAUGGCCACGGCAUUAUGUUCUUGUAUCUCGGUGCUCCAGCGUUGGAAUCAUCACAAGUUGAUUAA ..((((((..(((.((((((.....((((((.((((((((.(((((....((....))....))))).)))))))).)))))))))))..).)))..)).))))........ ( -38.50) >DroYak_CAF1 58265 112 + 1 UUCUUGGACGGUUGCUGCUGAUUGUGGACAUGGAGAUGCAUGGAUAAAUGGACACGGCAUUAUGUUCUUGUAUCUCGGUGCUCCACCGCUGUAAUAAUCACAAGUUGAUUAA ..((((((..(((((.((.....((((((((.((((((((.(((((((((.......)))).))))).)))))))).))).))))).)).)))))..)).))))........ ( -38.30) >DroMoj_CAF1 75646 104 + 1 CUCCCACAAUCUGAUUGCCUUUUGUGGAUGUACG--UAUGUGUGUAGAU--GCGCUGCAGUC----UUUCAAUCUUUGCGCUUCAUUGUUGUAAUCAUCAUAAAUUUAUUAA ...........(((((((.....(((.((.((((--......)))).))--.))).((((..----.........))))...........)))))))............... ( -16.20) >consensus UUCUUAGACUGUUGCUGCUGAUUGUGGGCAUGGAGAUGCAUGGAUAAAUGGCCACGGCAUUAUGUUAUUGUAUCUCGGUGCUCCAUCGUUGUAAUCAUCACAAGUUGAUUAA ......((..(((((.((....((.((((((.((((((((............................)))))))).))))))))..)).)))))..))............. (-15.69 = -17.17 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:42 2006