| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,766,944 – 6,767,054 |
| Length | 110 |
| Max. P | 0.978339 |

| Location | 6,766,944 – 6,767,054 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.04 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
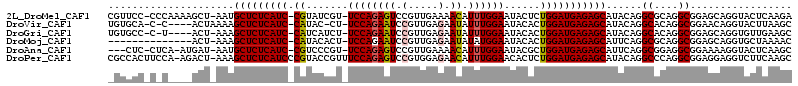
>2L_DroMel_CAF1 6766944 110 + 22407834 UCUUGAGUACCUGCUCCGCCUGCGCCUGUAUGCUCUCAUCCAGAGUAUUCCAAAUGUUUUCAACGGACUCUGGA-ACGAUACG-GAUGAGAGCAUU-AGCUUUUGGG-GGAACG ............(.(((.((.....(((.((((((((((((......(((((...((((.....))))..))))-)......)-))))))))))))-)).....)).-))).). ( -36.10) >DroVir_CAF1 46097 105 + 1 GCUUAAGUACCUGUUCCGCCUGUGCCUGUAUGCUCUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGGA-AG-GUAUG-GAUGAGAGCUUUUAGU----G-G-UGCACA ......(((((......((....))(((...(((((((((((.....(((((...((((.....))))..))))-).-...))-)))))))))...))).----)-)-)))... ( -36.60) >DroGri_CAF1 44299 105 + 1 GCUUCAACACCUGCUCCGCCUGUGCCUGUAUGCUCUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGAUUCUGGA-AGAUGAUG-GAUGAGAGCUUU-AGU----A-G-GGCACA ((..........)).......((((((....(((((((((((.....(((((...((((.....))))..))))-).....))-)))))))))...-...----.-)-))))). ( -39.00) >DroMoj_CAF1 44270 97 + 1 GUUUUAGCACCUGCUCCGCCUGCGCCUGAAUGCUCUCAUCCAGUGUAUUCCAUAUAUUCUCAACGGAUUCUGGA-AGUGUAUG-GAUGAGAGCUUU-AGU-------------- ......(((...((...)).)))..(((((.(((((((((((.....(((((...((((.....))))..))))-).....))-))))))))))))-)).-------------- ( -33.30) >DroAna_CAF1 33550 106 + 1 GCUUGAGUACCUUUUCCGCCUCCGCCUGAAUGCUCUCAUCCAGCGUAUUCCAAAUGUUUUCAACGGACUCUGGA-ACGGGACG-GAUGAGAGCAUU-AUCAU-UGAG-GAG--- ..................((((......(((((((((((((..(.(.(((((...((((.....))))..))))-).).)..)-))))))))))))-.....-.)))-)..--- ( -35.90) >DroPer_CAF1 38064 112 + 1 GCUUGAAGACCUCCUCCGCCUGGGCCUGUAUGCUCUCAUCCAGAGUGUUCCAAAUGUUCUCCACGGACUCUGGAAACGGUACGGGAUGAGAGCUUU-AGUCU-UGGAAGUGGCG ...............(((((..((.(((...((((((((((..(.(((((((...((((.....))))..))).)))).)...))))))))))..)-)).))-..)..)))).. ( -39.50) >consensus GCUUGAGUACCUGCUCCGCCUGCGCCUGUAUGCUCUCAUCCAGUGUAUUCCAAAUAUUCUCAACGGACUCUGGA_ACGGUACG_GAUGAGAGCUUU_AGU____G_G_GGCAC_ .................((....))......((((((((((.......((((...((((.....))))..))))........).)))))))))..................... (-20.76 = -20.04 + -0.72)

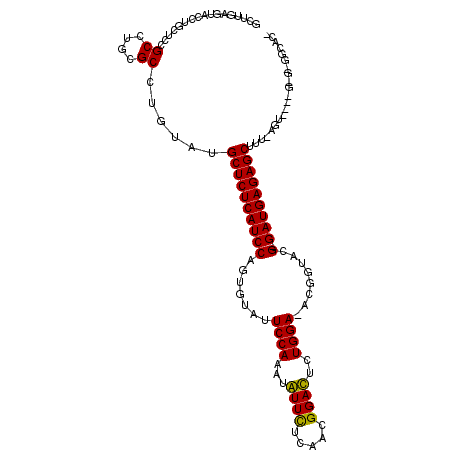

| Location | 6,766,944 – 6,767,054 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.54 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
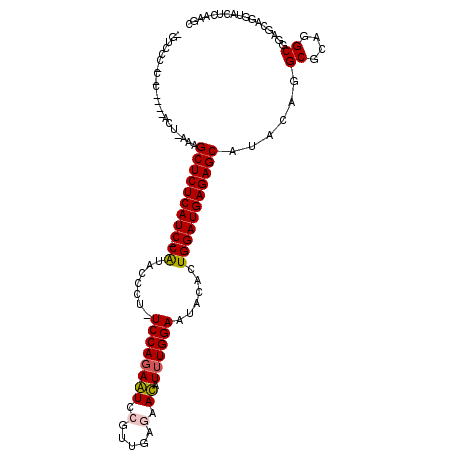
>2L_DroMel_CAF1 6766944 110 - 22407834 CGUUCC-CCCAAAAGCU-AAUGCUCUCAUC-CGUAUCGU-UCCAGAGUCCGUUGAAAACAUUUGGAAUACUCUGGAUGAGAGCAUACAGGCGCAGGCGGAGCAGGUACUCAAGA .(((((-.((....(((-.(((((((((((-((....((-((((((.((....)).....))))))))....)))))))))))))...)))...)).)))))............ ( -42.00) >DroVir_CAF1 46097 105 - 1 UGUGCA-C-C----ACUAAAAGCUCUCAUC-CAUAC-CU-UCCAGAAUCCGUUGAGAAUAUUUGGAAUACACUGGAUGAGAGCAUACAGGCACAGGCGGAACAGGUACUUAAGC .((((.-(-(----.......(((((((((-((...-.(-((((((.((......))...))))))).....)))))))))))......((....)))).....))))...... ( -31.10) >DroGri_CAF1 44299 105 - 1 UGUGCC-C-U----ACU-AAAGCUCUCAUC-CAUCAUCU-UCCAGAAUCCGUUGAGAAUAUUUGGAAUACACUGGAUGAGAGCAUACAGGCACAGGCGGAGCAGGUGUUGAAGC ((((((-.-.----...-...(((((((((-((.....(-((((((.((......))...))))))).....))))))))))).....)))))).((..(((....)))...)) ( -33.92) >DroMoj_CAF1 44270 97 - 1 --------------ACU-AAAGCUCUCAUC-CAUACACU-UCCAGAAUCCGUUGAGAAUAUAUGGAAUACACUGGAUGAGAGCAUUCAGGCGCAGGCGGAGCAGGUGCUAAAAC --------------...-...(((((((((-((.....(-((((.(.((......)).)...))))).....))))))))))).....(((((..((...))..)))))..... ( -32.80) >DroAna_CAF1 33550 106 - 1 ---CUC-CUCA-AUGAU-AAUGCUCUCAUC-CGUCCCGU-UCCAGAGUCCGUUGAAAACAUUUGGAAUACGCUGGAUGAGAGCAUUCAGGCGGAGGCGGAAAAGGUACUCAAGC ---(.(-(((.-.(((.-.(((((((((((-((.(..((-((((((.((....)).....))))))))..).))))))))))))))))....)))).)................ ( -37.30) >DroPer_CAF1 38064 112 - 1 CGCCACUUCCA-AGACU-AAAGCUCUCAUCCCGUACCGUUUCCAGAGUCCGUGGAGAACAUUUGGAACACUCUGGAUGAGAGCAUACAGGCCCAGGCGGAGGAGGUCUUCAAGC .(((.(((((.-.....-...((((((((((.(....(((((((((.((......))...))))))).)).).))))))))))......((....))))))).)))........ ( -39.70) >consensus _GUCCC_C_C____ACU_AAAGCUCUCAUC_CAUACCCU_UCCAGAAUCCGUUGAGAACAUUUGGAAUACACUGGAUGAGAGCAUACAGGCGCAGGCGGAGCAGGUACUCAAGC .....................(((((((((.((.......((((((((.(.....).)).))))))......)))))))))))......((....))................. (-21.79 = -21.54 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:36 2006