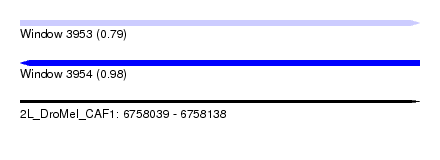
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,758,039 – 6,758,138 |
| Length | 99 |
| Max. P | 0.981253 |

| Location | 6,758,039 – 6,758,138 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.29 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -21.00 |
| Energy contribution | -20.68 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.58 |
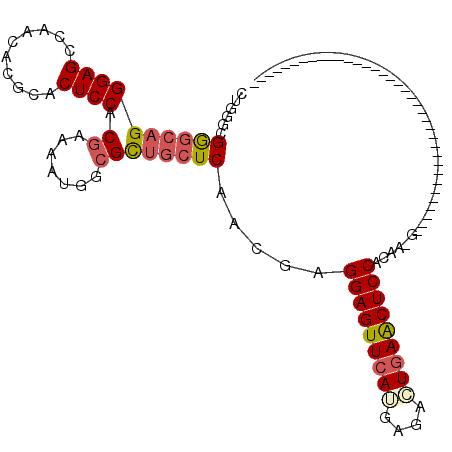
| SVM RNA-class probability | 0.789492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6758039 99 + 22407834 AGUCGCUGC------AUUCU------A------AUCCAUUGUGGAGUUCAGUCUCAUGAACUCCUCGUUGAGCAACGCCAUAUUCGUGGAGUGCGUGUUGGCUCCCUGCUCGCCCAG .......((------.....------.------.......(.((((((((......)))))))).)...(((((..((((....(((.....)))...))))....))))))).... ( -29.50) >DroSec_CAF1 24768 79 + 1 --------------------------------------UUGUGGAGUUCAGUCUCCUGAACUCCUCGUUGAGCAGCGCCAUUUUCGAGGAGUGCGUGUUGGCUCCCUGCCCGCCCAG --------------------------------------..(.(((((((((....))))))))).)..((.((((((((((((.....))))).)))))(((.....))).)).)). ( -29.30) >DroSim_CAF1 25503 79 + 1 --------------------------------------UUGUGGAGUUCAGUCUCAUGAACUCCUCGUUGAGCAGCGCCAUUUUCGCGGAGUGCGUGUUGGCUCCCUGCCCGCCCAG --------------------------------------..(.((((((((......)))))))).)((.(.((((.((((....((((.....)))).))))...))))).)).... ( -28.30) >DroEre_CAF1 28346 84 + 1 ---------------------------------GCCCGUGGUGGAGCUUAUCUUCAUGAACUCCUCGUUGAGCAGCGCCAUCUUCGUGGAGUGCGUGGUGCCUCCCAUGCCUCCAUG ---------------------------------......(((((.(((...(((.((((.....)))).))).))).)))))..(((((((.((((((......))))))))))))) ( -33.00) >DroYak_CAF1 27154 117 + 1 AGUCGCUCCCCGUGGAGUUUAGCUUUAAGAACUCCUCGUCGUGGAGUUCAUCUGGAUGAACUCCCGGUUGAGCAGCGUCAUCUUGGAGGAGUGCAUGGUGCCUCCCUGCUCGCCCAG ....((...(((.((((((((.((....(((((((.......)))))))....)).)))))))))))..((((((..((......))((((.((.....)))))))))))))).... ( -45.50) >consensus ____________________________________C_UUGUGGAGUUCAGUCUCAUGAACUCCUCGUUGAGCAGCGCCAUCUUCGAGGAGUGCGUGUUGGCUCCCUGCCCGCCCAG ..........................................(((((((((....))))))))).....(((((((((((.(((....))))).)))))).))).(((......))) (-21.00 = -20.68 + -0.32)

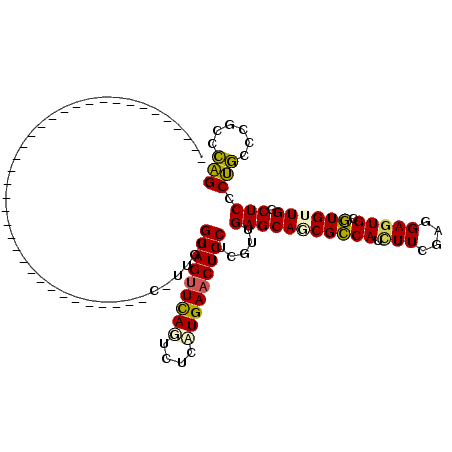
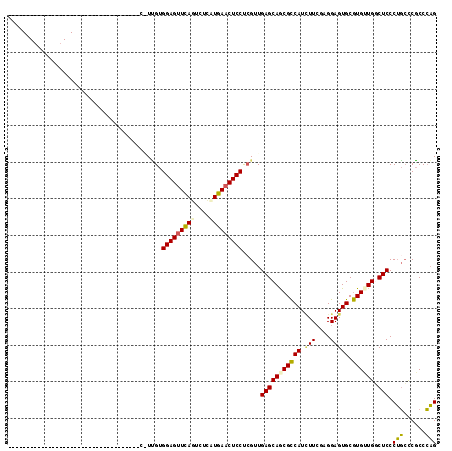
| Location | 6,758,039 – 6,758,138 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.29 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -22.60 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6758039 99 - 22407834 CUGGGCGAGCAGGGAGCCAACACGCACUCCACGAAUAUGGCGUUGCUCAACGAGGAGUUCAUGAGACUGAACUCCACAAUGGAU------U------AGAAU------GCAGCGACU ....((((((((...((((...((.......))....)))).)))))).....((((((((......)))))))).........------.------.....------...)).... ( -27.90) >DroSec_CAF1 24768 79 - 1 CUGGGCGGGCAGGGAGCCAACACGCACUCCUCGAAAAUGGCGCUGCUCAACGAGGAGUUCAGGAGACUGAACUCCACAA-------------------------------------- .(((((((((.(((((..........)))))........)).)))))))....(((((((((....)))))))))....-------------------------------------- ( -34.60) >DroSim_CAF1 25503 79 - 1 CUGGGCGGGCAGGGAGCCAACACGCACUCCGCGAAAAUGGCGCUGCUCAACGAGGAGUUCAUGAGACUGAACUCCACAA-------------------------------------- ......((((((...((((...(((.....)))....)))).)))))).....((((((((......))))))))....-------------------------------------- ( -30.90) >DroEre_CAF1 28346 84 - 1 CAUGGAGGCAUGGGAGGCACCACGCACUCCACGAAGAUGGCGCUGCUCAACGAGGAGUUCAUGAAGAUAAGCUCCACCACGGGC--------------------------------- ..(((((((.(((......))).)).))))).............((((...(.((((((.((....)).)))))).)...))))--------------------------------- ( -24.40) >DroYak_CAF1 27154 117 - 1 CUGGGCGAGCAGGGAGGCACCAUGCACUCCUCCAAGAUGACGCUGCUCAACCGGGAGUUCAUCCAGAUGAACUCCACGACGAGGAGUUCUUAAAGCUAAACUCCACGGGGAGCGACU ....((((((((((((((.....)).))))((......))..))))))..((((((((((((....))))))))).......((((((..........)))))).)))...)).... ( -44.40) >consensus CUGGGCGGGCAGGGAGCCAACACGCACUCCACGAAAAUGGCGCUGCUCAACGAGGAGUUCAUGAGACUGAACUCCACAA_G____________________________________ ......((((((((((..........)))).((.......)))))))).....(((((((((....))))))))).......................................... (-22.60 = -23.28 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:34 2006