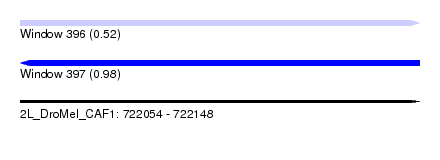
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 722,054 – 722,148 |
| Length | 94 |
| Max. P | 0.980470 |

| Location | 722,054 – 722,148 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 722054 94 + 22407834 GAAGAUCACCGAAGGUAACCAGGGAAUGCUUAUUGGAGCCACAUGCCAGUGCACUGGCAAAAAUAGUCGUGUUGGGC-AUCGGACUCCAAUUGGU .....((.((...((...)).))))..(((.(((((((((..(((((...((((.(((.......)))))))..)))-)).)).))))))).))) ( -29.90) >DroSec_CAF1 100677 92 + 1 --AGAUCACCGAAGGUAACGAGGGAAUGCUUAUUGGAGCCACAUGCCAGUGCACUGGCAGAAAUAGUCGUGUUGGGC-CUCGGACUCCAAUUGAA --......((((.(((.((((......((((....))))....((((((....)))))).......)))).....))-)))))............ ( -26.00) >DroSim_CAF1 105813 92 + 1 --AGAUCACCGAAGGUAACCAGGGAAUGCUUAUUGGAGCCACAUGCCAGUGCACUGGCAAAAAUAGUCGUGUUGGGC-CUCGGACUCCAAUUGGA --......(((((((((.(....)..))))).((((((((...((((((....))))))......(((......)))-...)).)))))))))). ( -27.00) >DroEre_CAF1 111516 95 + 1 ACGGAUCACCGAAGGUAACCAGGGAAUGCUUAUUGGAGCUACAUGCCAGUGCACUGGCAAAAAUAGUCGUGUUGGGCUCUGAGACUCCAAUUGGA .(((....))).................((.(((((((((....(((...((((.(((.......)))))))..)))....)).))))))).)). ( -27.40) >DroYak_CAF1 106781 94 + 1 AAAGAUCACCGAAGGUAACCAGGGAAUGCUUAUUGGAGCUACAUGCCAGUGCACUGGCAAAAAUAGUCGUGUUGGGC-CUCGGACUCCAAUUGGA ........((((.(((..((((.(...((((....))))....((((((....))))))..........).))))))-)))))..(((....))) ( -27.20) >consensus __AGAUCACCGAAGGUAACCAGGGAAUGCUUAUUGGAGCCACAUGCCAGUGCACUGGCAAAAAUAGUCGUGUUGGGC_CUCGGACUCCAAUUGGA .....((.((...(....)..))))...((.(((((((((....(((...((((.(((.......)))))))..)))....)).))))))).)). (-25.24 = -25.04 + -0.20)

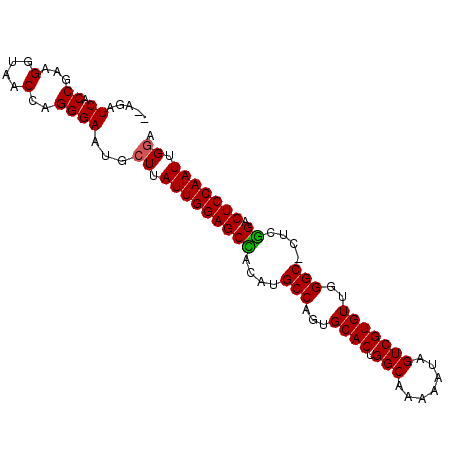
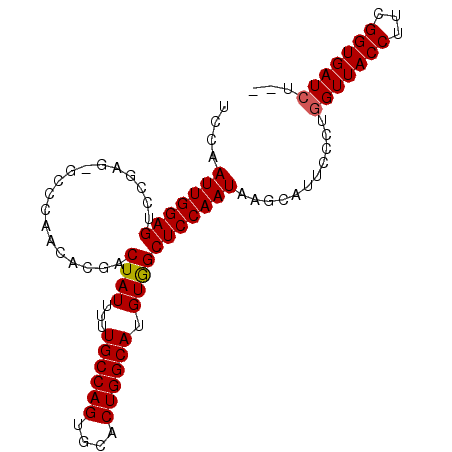
| Location | 722,054 – 722,148 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.42 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 722054 94 - 22407834 ACCAAUUGGAGUCCGAU-GCCCAACACGACUAUUUUUGCCAGUGCACUGGCAUGUGGCUCCAAUAAGCAUUCCCUGGUUACCUUCGGUGAUCUUC ....(((((((((((.(-(.....))))........((((((....))))))...)))))))))...........(((((((...)))))))... ( -28.20) >DroSec_CAF1 100677 92 - 1 UUCAAUUGGAGUCCGAG-GCCCAACACGACUAUUUCUGCCAGUGCACUGGCAUGUGGCUCCAAUAAGCAUUCCCUCGUUACCUUCGGUGAUCU-- ....(((((((((((.(-......).))........((((((....))))))...)))))))))............((((((...))))))..-- ( -25.90) >DroSim_CAF1 105813 92 - 1 UCCAAUUGGAGUCCGAG-GCCCAACACGACUAUUUUUGCCAGUGCACUGGCAUGUGGCUCCAAUAAGCAUUCCCUGGUUACCUUCGGUGAUCU-- ....(((((((((((.(-......).))........((((((....))))))...)))))))))...........(((((((...))))))).-- ( -28.80) >DroEre_CAF1 111516 95 - 1 UCCAAUUGGAGUCUCAGAGCCCAACACGACUAUUUUUGCCAGUGCACUGGCAUGUAGCUCCAAUAAGCAUUCCCUGGUUACCUUCGGUGAUCCGU ....((((((((..(((....)..............((((((....))))))))..))))))))...........(((((((...)))))))... ( -25.50) >DroYak_CAF1 106781 94 - 1 UCCAAUUGGAGUCCGAG-GCCCAACACGACUAUUUUUGCCAGUGCACUGGCAUGUAGCUCCAAUAAGCAUUCCCUGGUUACCUUCGGUGAUCUUU ....(((((((..((.(-......).)).((((...((((((....)))))).)))))))))))...........(((((((...)))))))... ( -27.50) >consensus UCCAAUUGGAGUCCGAG_GCCCAACACGACUAUUUUUGCCAGUGCACUGGCAUGUGGCUCCAAUAAGCAUUCCCUGGUUACCUUCGGUGAUCU__ ....(((((((..................((((...((((((....)))))).)))))))))))...........(((((((...)))))))... (-23.46 = -23.42 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:08 2006