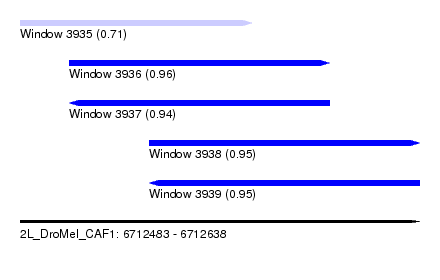
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,712,483 – 6,712,638 |
| Length | 155 |
| Max. P | 0.958306 |

| Location | 6,712,483 – 6,712,573 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -28.54 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
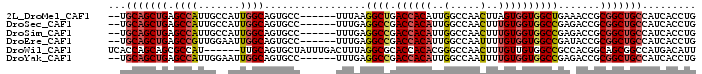
>2L_DroMel_CAF1 6712483 90 + 22407834 --UGCAGCUGAGCCAUUGCCAUUGGCAGUGCC------UUUAAGGCUGACCACAUUGGCCAACUUAGUGGUGGCUGAAACCGCGGCUGCCAUCACCUG --.(((((((((.(((((((...))))))).)------))...(((..(.....)..)))......(((((.......)))))))))))......... ( -32.80) >DroSec_CAF1 25107 90 + 1 --UGCAGCUGAGCCAUUGCCAUUGGCAGUGCC------UUUGAGGCCGACCACAUUGGCCAACUUUGUGGUGGCCGAGACCGCGGCUGCCAUCACCUG --.(((((((((.(((((((...))))))).)------)....((((.((((((..(.....)..)))))))))).......)))))))......... ( -39.70) >DroSim_CAF1 24973 90 + 1 --UGCAGCUGAGCCAUUGCCAUUGGCAGUGCC------UUUGAGGCCGACCACAUUGGCCAACUUUGUGGUGGCCGAGACCGCGGCUGCCAUCACCUG --.(((((((((.(((((((...))))))).)------)....((((.((((((..(.....)..)))))))))).......)))))))......... ( -39.70) >DroEre_CAF1 26340 90 + 1 --UGCAGCUGAGCCGUUGGAAUUGGCAGUGCC------UUUGAGGCCGACCACAUUGGCCAAUUUUGUGGUGGCCGAUACCGCGGCUGCCAUCACCUG --.(((((((.((((.......))))......------.....((((.((((((...........)))))))))).......)))))))......... ( -33.80) >DroWil_CAF1 43693 92 + 1 UCACCAGCAGCGCCAU------UUGCAGUGCUAUUUGACUUUAGGCGCACCACACGGGCCAACUUUGUUGUGGCCGCCACGGCAGCGGCCAUGACAUU ......((.(((((..------..((((......))).)....))))).((....))))......(((..(((((((.......)))))))..))).. ( -33.40) >DroYak_CAF1 26614 90 + 1 --UGCAGCUGAGCCAUUGGAAUUGGCAGUGCC------UUUGAGGCCGACCACAUUGGCCAAUUUUGUGGUGGCCGAGACCGCGGCUGCCAUCACCUG --.((((((..((((.......))))...((.------.((..((((.((((((...........))))))))))..))..))))))))......... ( -34.90) >consensus __UGCAGCUGAGCCAUUGCCAUUGGCAGUGCC______UUUGAGGCCGACCACAUUGGCCAACUUUGUGGUGGCCGAGACCGCGGCUGCCAUCACCUG ...(((((((.((((.......)))).................((((.((((((..(.....)..)))))))))).......)))))))......... (-28.54 = -29.10 + 0.56)
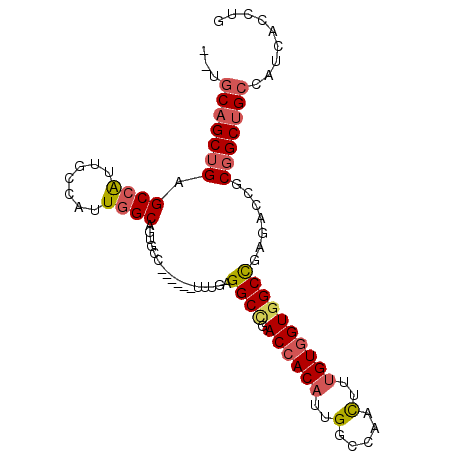
| Location | 6,712,502 – 6,712,603 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -21.57 |
| Energy contribution | -22.65 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6712502 101 + 22407834 UUGGCAGUGCC------UUUAAGGCUGACCACAUUGGCCAACUUAGUGGUGGCUGAAACCGCGGCUGCCAUCACCUGGCCUCUUCCAUUCAGCAGCGAGUUGUUGGU- .(((((..(((------.....))))).)))....(((((.....((((..((((......))))..))))....)))))........(((((((....))))))).- ( -35.40) >DroVir_CAF1 41015 106 + 1 CCGGCAGCACCAUUUGUCUUAAGGCGCACCACAUGCGCCAGCUUUGUGGUGGCCGCAACUGCAGCAGCCAUCACAUUGCCAUUGUUAUUGGGCAACGUGUUGAUGG-- (((.((((((...((((((...((((((.....)))))).((..(((((((((.((.......)).)))))))))..))..........)))))).)))))).)))-- ( -46.00) >DroPse_CAF1 42628 99 + 1 CC---GUUGCU------CUUGAGGCCCACAACAUGGGCCAACUUUGUUGUGGCCGCCACCGCAGCGGCCAUCACAGUGCCAUGUCUAUUGGCCAGAGAAUUGUUGUUG .(---(...((------((...((((((.....)))))).....(((.((((((((.......)))))))).)))(.((((.......)))))))))...))...... ( -37.60) >DroWil_CAF1 43709 105 + 1 -UUGCAGUGCUAUUUGACUUUAGGCGCACCACACGGGCCAACUUUGUUGUGGCCGCCACGGCAGCGGCCAUGACAUUGCCAUGACCGUUGGGUAAGGCAUUGUUGA-- -..(((((((((((..((.((((((((.((....))))......(((..(((((((.......)))))))..)))..))).)))..))..)))..))))))))...-- ( -41.90) >DroYak_CAF1 26633 101 + 1 UUGGCAGUGCC------UUUGAGGCCGACCACAUUGGCCAAUUUUGUGGUGGCCGAGACCGCGGCUGCCAUCACCUGGCCACGUCCAUUGAGCAGCGAGUUGUUGGU- ..(((...)))------......((((((.((..((((((.....((((..((((......))))..))))....))))))(((..........))).)).))))))- ( -37.70) >DroPer_CAF1 42021 100 + 1 CCG--GUUGCU------CUUGAGGCCCACAACAUGGGCCAACUUCGUUGUGGCCGCCACCGCAGCGGCCAUCACAGUGCCAUGUCUAUUGGCCAGAGAAUUGUUGUUG .((--(((.((------((...((((((.....))))))......((.((((((((.......)))))))).)).(.((((.......))))))))))))))...... ( -39.30) >consensus CCGGCAGUGCC______CUUGAGGCCCACCACAUGGGCCAACUUUGUGGUGGCCGCCACCGCAGCGGCCAUCACAUUGCCAUGUCCAUUGGGCAGCGAAUUGUUGGU_ ..(((.................((((((.....))))))......((((((((.((.......)).))))))))...)))............................ (-21.57 = -22.65 + 1.08)

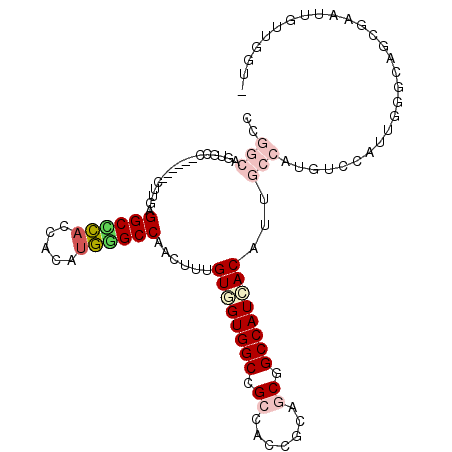
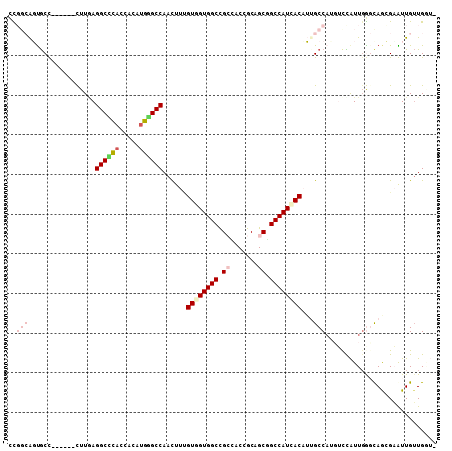
| Location | 6,712,502 – 6,712,603 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6712502 101 - 22407834 -ACCAACAACUCGCUGCUGAAUGGAAGAGGCCAGGUGAUGGCAGCCGCGGUUUCAGCCACCACUAAGUUGGCCAAUGUGGUCAGCCUUAAA------GGCACUGCCAA -....................(((.((..(((.((((.(((((((....)))...)))).))))..(((((((.....)))))))......------))).)).))). ( -30.90) >DroVir_CAF1 41015 106 - 1 --CCAUCAACACGUUGCCCAAUAACAAUGGCAAUGUGAUGGCUGCUGCAGUUGCGGCCACCACAAAGCUGGCGCAUGUGGUGCGCCUUAAGACAAAUGGUGCUGCCGG --((.....(((((((((..........)))))))))..(((.(((((....)))))(((((.....(((((((((...)))))))...)).....)))))..))))) ( -42.20) >DroPse_CAF1 42628 99 - 1 CAACAACAAUUCUCUGGCCAAUAGACAUGGCACUGUGAUGGCCGCUGCGGUGGCGGCCACAACAAAGUUGGCCCAUGUUGUGGGCCUCAAG------AGCAAC---GG ...........((((((((....(((((((((((.((.((((((((.....))))))))))....)))..).)))))))...))))...))------))....---.. ( -38.00) >DroWil_CAF1 43709 105 - 1 --UCAACAAUGCCUUACCCAACGGUCAUGGCAAUGUCAUGGCCGCUGCCGUGGCGGCCACAACAAAGUUGGCCCGUGUGGUGCGCCUAAAGUCAAAUAGCACUGCAA- --(((((..((((..(((....)))...)))).(((..((((((((.....))))))))..)))..)))))....((..((((...............))))..)).- ( -39.76) >DroYak_CAF1 26633 101 - 1 -ACCAACAACUCGCUGCUCAAUGGACGUGGCCAGGUGAUGGCAGCCGCGGUCUCGGCCACCACAAAAUUGGCCAAUGUGGUCGGCCUCAAA------GGCACUGCCAA -.....((.((.((..(((....)).)..)).)).)).((((((.....((((.((((((((((...........)))))).))))....)------))).)))))). ( -34.60) >DroPer_CAF1 42021 100 - 1 CAACAACAAUUCUCUGGCCAAUAGACAUGGCACUGUGAUGGCCGCUGCGGUGGCGGCCACAACGAAGUUGGCCCAUGUUGUGGGCCUCAAG------AGCAAC--CGG ...........((((((((....(((((((((((.((.((((((((.....))))))))))....)))..).)))))))...))))...))------))....--... ( -38.00) >consensus _ACCAACAACUCGCUGCCCAAUAGACAUGGCAAUGUGAUGGCCGCUGCGGUGGCGGCCACAACAAAGUUGGCCCAUGUGGUGGGCCUCAAG______AGCACUGCCAG .............................((..(((..(((((((.......)))))))..))).....((((((.....))))))............))........ (-21.01 = -21.68 + 0.67)
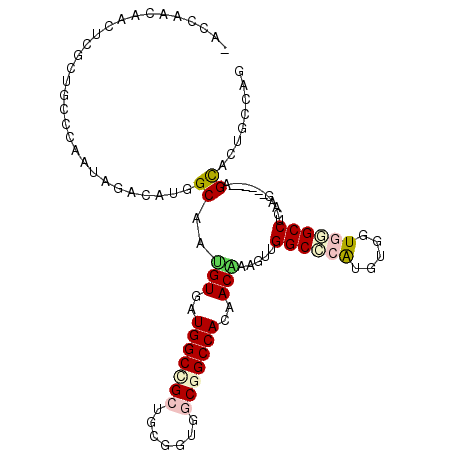

| Location | 6,712,533 – 6,712,638 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -17.57 |
| Energy contribution | -19.35 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6712533 105 + 22407834 CCAACUUAGUGGUGGCUGAAACCGCGGCUGCCAUCACCUGGCCUCUUCCAUUCAGCAGCG---AGUUGUUGGU------CAACUGCUUUUGCAUCAGCUGAUUGGAGGAGUCCA ........((((..((((......))))..)))).....((.((((((((.(((((....---(((((.....------)))))((....))....))))).)))))))).)). ( -44.90) >DroVir_CAF1 41052 99 + 1 CCAGCUUUGUGGUGGCCGCAACUGCAGCAGCCAUCACAUUGCCAUUGUUAUUGGGCAACG---UGUUGAUGG------------GCUUCUGCAGCAGCUGAUUGGAGGAGUCCA .((((..(((((((((.((.......)).)))))))))(((((..........)))))..---.)))).(((------------((((((.((((....).))).))))))))) ( -39.90) >DroPse_CAF1 42656 111 + 1 CCAACUUUGUUGUGGCCGCCACCGCAGCGGCCAUCACAGUGCCAUGUCUAUUGGCCAGAG---AAUUGUUGUUGGCCGCAGCCGGCUUCUGCAUGAGUUGAUUGGAAGAGUCCA .(((((((((...(((((.....((.(((((((.((((((((((.......)))).....---.)))).)).))))))).)))))))...))).))))))..((((....)))) ( -41.70) >DroWil_CAF1 43745 102 + 1 CCAACUUUGUUGUGGCCGCCACGGCAGCGGCCAUGACAUUGCCAUGACCGUUGGGUAAGG---CAUUGUUGA---------CUGGCUUCUGAAGCAGCUGAUUGGAUGAGUCCA .((((..(((..(((((((.......)))))))..))).((((...(((....)))..))---))..)))).---------..((((.(....).))))...((((....)))) ( -37.50) >DroMoj_CAF1 41136 99 + 1 CCAGCUUUGUGGUGGCAGCCACAGCAGCAGCCACCACAUUGCCAUUGCCAUUGGGCAGCG---UGUUGAUGG------------GCUUCUGCAGUAAUUGAUUGGAGGAGUCCA ...((..(((((((((.((.......)).)))))))))..))..(((((....)))))..---......(((------------((((((.((((.....)))).))))))))) ( -44.20) >DroAna_CAF1 29692 108 + 1 CCAACUUCGUUGUGGCUGCCACAGCCGCAGCCAUCACCUGGCCAUGUCCGUUAGCCAUCGAGGAGUUAUUGGC------AGGCGGCUUUUGCACCAGCUGGUUGGAGGAGUCCA ...(((((((((((((((...))))))))))......(..((((.(.((((..((((............))))------..)))))....((....))))))..).)))))... ( -40.60) >consensus CCAACUUUGUGGUGGCCGCCACAGCAGCAGCCAUCACAUUGCCAUGUCCAUUGGGCAGCG___AGUUGUUGG__________CGGCUUCUGCAGCAGCUGAUUGGAGGAGUCCA (((((...((((((((.((.......)).))))))))...(((..........)))...........)))))............((((((.((.........)).))))))... (-17.57 = -19.35 + 1.78)

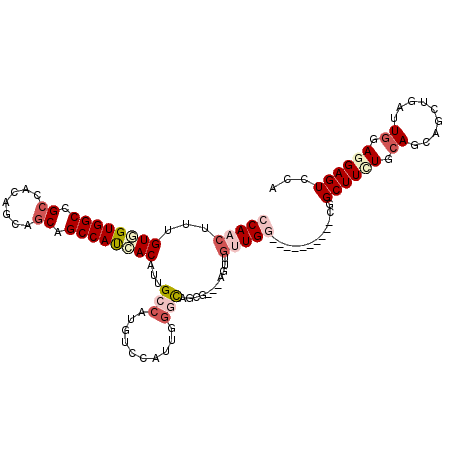
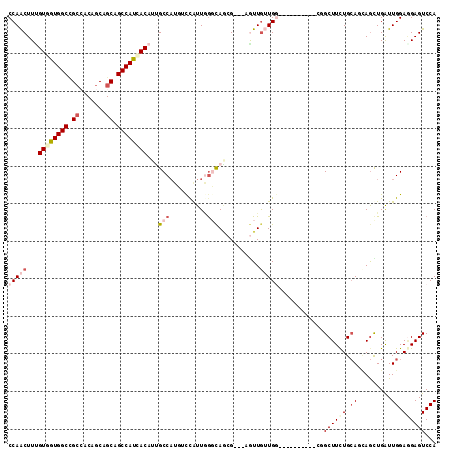
| Location | 6,712,533 – 6,712,638 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6712533 105 - 22407834 UGGACUCCUCCAAUCAGCUGAUGCAAAAGCAGUUG------ACCAACAACU---CGCUGCUGAAUGGAAGAGGCCAGGUGAUGGCAGCCGCGGUUUCAGCCACCACUAAGUUGG (((.(((.((((.(((((.........((((((((------.....)))))---.)))))))).)))).))).)))((((.(((((((....)))...)))).))))....... ( -37.40) >DroVir_CAF1 41052 99 - 1 UGGACUCCUCCAAUCAGCUGCUGCAGAAGC------------CCAUCAACA---CGUUGCCCAAUAACAAUGGCAAUGUGAUGGCUGCUGCAGUUGCGGCCACCACAAAGCUGG ((((....))))..((((((((((((.(((------------(.(((...(---(((((((..........))))))))))))))).)))))))((.((...)).)).))))). ( -39.20) >DroPse_CAF1 42656 111 - 1 UGGACUCUUCCAAUCAACUCAUGCAGAAGCCGGCUGCGGCCAACAACAAUU---CUCUGGCCAAUAGACAUGGCACUGUGAUGGCCGCUGCGGUGGCGGCCACAACAAAGUUGG ((((....))))..(((((.........((((.(((.(((((.........---...)))))..)))...))))..(((..((((((((.....))))))))..))).))))). ( -39.70) >DroWil_CAF1 43745 102 - 1 UGGACUCAUCCAAUCAGCUGCUUCAGAAGCCAG---------UCAACAAUG---CCUUACCCAACGGUCAUGGCAAUGUCAUGGCCGCUGCCGUGGCGGCCACAACAAAGUUGG ((((....))))....((((((.....)).)))---------)((((..((---((..(((....)))...)))).(((..((((((((.....))))))))..)))..)))). ( -35.30) >DroMoj_CAF1 41136 99 - 1 UGGACUCCUCCAAUCAAUUACUGCAGAAGC------------CCAUCAACA---CGCUGCCCAAUGGCAAUGGCAAUGUGGUGGCUGCUGCUGUGGCUGCCACCACAAAGCUGG ((((....))))..........((....))------------(((......---((.((((....)))).))((..(((((((((.(((.....))).)))))))))..))))) ( -33.10) >DroAna_CAF1 29692 108 - 1 UGGACUCCUCCAACCAGCUGGUGCAAAAGCCGCCU------GCCAAUAACUCCUCGAUGGCUAACGGACAUGGCCAGGUGAUGGCUGCGGCUGUGGCAGCCACAACGAAGUUGG ((((....)))).(((((((((((...((((((..------((((...(((......((((((.......)))))))))..)))).))))))...))).)).......)))))) ( -40.11) >consensus UGGACUCCUCCAAUCAGCUGCUGCAGAAGCCG__________CCAACAACU___CGCUGCCCAAUGGAAAUGGCAAUGUGAUGGCUGCUGCGGUGGCGGCCACAACAAAGUUGG ((((....)))).((((((...((....))..............................................(((..(((((((.......)))))))..))).)))))) (-19.13 = -19.22 + 0.09)

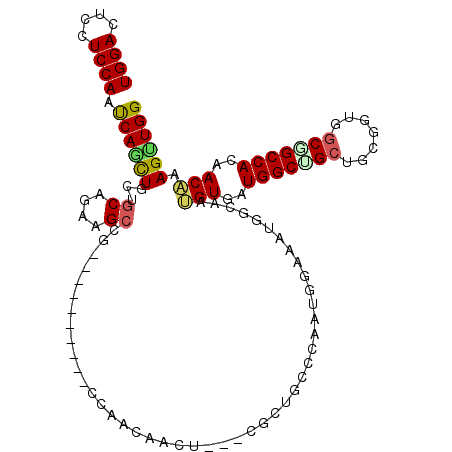
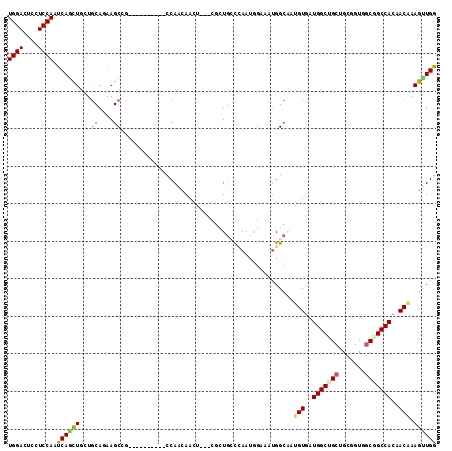
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:20 2006