
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,707,753 – 6,707,867 |
| Length | 114 |
| Max. P | 0.844092 |

| Location | 6,707,753 – 6,707,867 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.45 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -13.83 |
| Energy contribution | -16.67 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
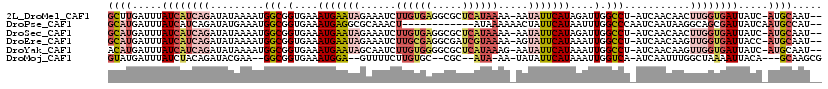
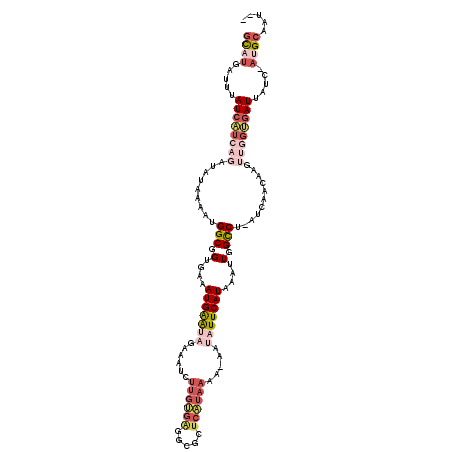
>2L_DroMel_CAF1 6707753 114 + 22407834 GCUUGAUUUAUCAUCAGAUAUAAAAUGGCGGUGAAAUGAAUAGAAAUCUUGUGAGGCGCUCAUAAAA-AAUAUUCAUAGAUUGGCCU-AUCAACAACUUGGUGAUUAUC-AUGCAAU-- ((.((((..((((((((.........(((.(....(((((((......(((((((...)))))))..-..)))))))....).))).-.........)))))))).)))-).))...-- ( -28.41) >DroPse_CAF1 34747 105 + 1 GCAUGAUUUAUCAUCAGAUAUGAAAUGGCGGUGAAAUGAGGCGCAAACU------------AUAAAAAACUAUUCAUAAUUUGGCCCAAUCAAUAAGGCAGCGAUUAUCAAUGCCAU-- (((((((..(((.(((....)))..((.(..(((..((.(((.((((.(------------((.((......)).))).))))))))).)))....).))..))).))).))))...-- ( -18.40) >DroSec_CAF1 20480 114 + 1 GCAUGAUUUAUCAUCAGAUAUAAAAUGGCGGUGAAAUGAAUAGAAAUCUUGUGAGGCGCUCAUAAAA-AAUAUUCAUAGAUUGGCCU-AUCAACAACUUGGUGAUUAUC-AUGCAAU-- (((((((..((((((((.........(((.(....(((((((......(((((((...)))))))..-..)))))))....).))).-.........)))))))).)))-))))...-- ( -32.71) >DroEre_CAF1 21451 114 + 1 GCAUGAUUUAUCAUCAGAUAUAAAAUGGCGGUGAAAUGAAUAGAAAUCUUGCGAGGCGAUCGUAAAA-AGUAUUCAUAAAUUGGCCU-AUCAACAAGUUGGUGAUUACC-AUGCAAU-- (((((....((((((((.........(((.(....(((((((......((((((.....))))))..-..)))))))....).))).-.........))))))))...)-))))...-- ( -29.71) >DroYak_CAF1 21366 114 + 1 ACAUGAUUUAUCAUCAGAUAUAAAAUGGCGGUGAAAUGAAUAGCAAUCUUGUGGGGCGCUCAUAAAG-AAUAUUCAUAAAUUGGCCU-AUCAACAAGUUGGUGAUUAUC-AUGCAAU-- .((((((..((((((((.........(((.(....(((((((....(((((((((...))))).)))-).)))))))....).))).-.........)))))))).)))-)))....-- ( -29.61) >DroMoj_CAF1 33877 105 + 1 GUAUGAUUUAUCUACAGAUACGAA--GGCGGUGAAAUGGA--GUUUUCUUGUGC--CGC--AUA-AA-UAUAUUCAUAAAUUGGUCA-AUCAAUUUGGCUAAAAUUACA---GCAAGCG .(((((..((((....))))....--.((((((....((.--.....))..)))--)))--...-..-.....)))))..(((((((-(.....)))))))).......---....... ( -20.50) >consensus GCAUGAUUUAUCAUCAGAUAUAAAAUGGCGGUGAAAUGAAUAGAAAUCUUGUGAGGCGCUCAUAAAA_AAUAUUCAUAAAUUGGCCU_AUCAACAAGUUGGUGAUUAUC_AUGCAAU__ ((((.....((((((((.........(((.(....(((((((......((((((.....)))))).....)))))))....).)))...........)))))))).....))))..... (-13.83 = -16.67 + 2.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:12 2006