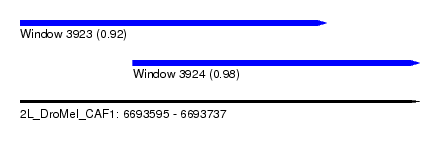
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,693,595 – 6,693,737 |
| Length | 142 |
| Max. P | 0.980215 |

| Location | 6,693,595 – 6,693,704 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6693595 109 + 22407834 AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGCGAGGUUG-----AGGUUGGGGC-UCCAGCUCCAGCUCCAUCUCCAACUCCAACUUCAACUCCUUCUCUAUCUACAACUC ..((((((.......))))..((....)).....))(((((((-----..(((((((.-...(((....)))....)))))))..)))))))....................... ( -31.10) >DroVir_CAF1 17607 100 + 1 AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGUGAGGUUGGCAGUAGGUUG--GCUGCCGGCUCCAGCUUCAGCUUCAGCUUUAGCCGCGACUCGAUCU------------- ..(((((.........((((((((.......)))))).((((((((((......--))))))))))...))...(((....)))...)))))..........------------- ( -34.80) >DroSec_CAF1 6612 103 + 1 AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGCGAGGUUG-----AGGUUGGGGC-UCCAGCUCCAGCUCCA------GCUCAAACUUCAACUCCUUCUCCAUCUACAACUC ..((((((.......))))..((....)).....))(..((((-----(((((.((((-(..(((....)))..)------)))).)))))))))..)................. ( -29.80) >DroSim_CAF1 6540 103 + 1 AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGCGAGGUUG-----AGGUUGGGGC-UCCAGCUCCAGCUCCA------GCUCCAACUUCAACUCCUUCUCCAUCUACAACUC ..((((((.......))))..((....)).....))(..((((-----((((((((((-(..(((....)))..)------))))))))))))))..)................. ( -36.30) >DroEre_CAF1 6486 91 + 1 AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGCGAGGUUG-----AGGUUGGGGC-UCCAGCUCCAGCUCCA------ACUCCAGCUC------------CAUCUCCAACUC ..(.((((.......)))).).((........(((.(((.(((-----..((((((((-....))))))))..))------)))))))...------------.....))..... ( -30.59) >DroYak_CAF1 6467 103 + 1 AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGCGAGGUUC-----AGGCUGGGGA-UCCAACUCCAGCUCCA------GCUCCAUCUCCAUCUCCGACUCCAUCUCCAGCUC .(((((((.......))))...((.........((.(((((..-----.((((((((.-...........)))))------)))..))))))).........)).......))). ( -25.87) >consensus AAGCGGCGUUUAUUUUGCCACAGGCCACUUAUCUGCGAGGUUG_____AGGUUGGGGC_UCCAGCUCCAGCUCCA______GCUCCAACUUCAACUCCUUCUCCAUCUACAACUC ..((((((.......))))..((....)).....))(((..........(((((((((.....)))))))))..........))).............................. (-18.78 = -18.87 + 0.09)

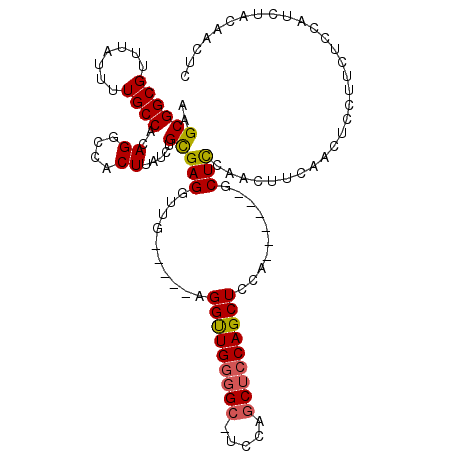

| Location | 6,693,635 – 6,693,737 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.98 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.12 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6693635 102 + 22407834 UUGAGGUUGGGGCUCCAGCUCCAGCUCCAUCUCCAACUCCAACUUCAACUCCUUCUCUAUCUACAACUCCGACUCCGGCUGCAGCUUUUCAGCUCUUUGGCU .((..((((((((....))))))))..)).......................................((((....(((((........)))))..)))).. ( -22.30) >DroSec_CAF1 6652 96 + 1 UUGAGGUUGGGGCUCCAGCUCCAGCUCCA------GCUCAAACUUCAACUCCUUCUCCAUCUACAACUCCGACUCCGGCUGCAGCUUUUCAGCUCUUUGGCU ((((((((.(((((..(((....)))..)------)))).))))))))....................((((....(((((........)))))..)))).. ( -24.70) >DroSim_CAF1 6580 96 + 1 UUGAGGUUGGGGCUCCAGCUCCAGCUCCA------GCUCCAACUUCAACUCCUUCUCCAUCUACAACUCCGACUCCGGCUGCAGCUUUUCAGCUCUUUGGCU ((((((((((((((..(((....)))..)------)))))))))))))....................((((....(((((........)))))..)))).. ( -31.20) >DroEre_CAF1 6526 84 + 1 UUGAGGUUGGGGCUCCAGCUCCAGCUCCA------ACUCCAGCUC------------CAUCUCCAACUCCGACUCCGGCUGCAGCUUUUCAGCUCUUUGGCU (((..(((((((((........)))))))------))..)))...------------...........((((....(((((........)))))..)))).. ( -23.10) >DroYak_CAF1 6507 96 + 1 UUCAGGCUGGGGAUCCAACUCCAGCUCCA------GCUCCAUCUCCAUCUCCGACUCCAUCUCCAGCUCCAACUCCGGCUGCAGCUUUUCAGCUCUUUGGCU ....((((((((((........(((....------)))..............(....)))))))))))((((....(((((........)))))..)))).. ( -21.70) >consensus UUGAGGUUGGGGCUCCAGCUCCAGCUCCA______GCUCCAACUUCAACUCCUUCUCCAUCUACAACUCCGACUCCGGCUGCAGCUUUUCAGCUCUUUGGCU .((.(((((((((....))))))))).)).......................................((((....(((((........)))))..)))).. (-20.12 = -20.20 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:07 2006