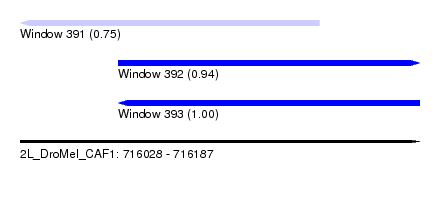
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 716,028 – 716,187 |
| Length | 159 |
| Max. P | 0.998511 |

| Location | 716,028 – 716,147 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -28.26 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 716028 119 - 22407834 GUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGUCGCGAUUAAAACGGCAAGAGUCAAAAAGUUGAAGAACGAGAAAGCGCACUUCAAAUGCCAUUUCAAUGGCGUCAAAUUGU- (((((((((.....(((.((((.((..(((((((.......)))))))..)))))).))).......))))))))).................(((((((....)))))))........- ( -33.40) >DroSec_CAF1 94802 119 - 1 GUUCUUCAGAUCUUGCUAUUGCACGGGUCGAUUGGCAGCCGCGAUUAAAACGGCAAGAGUCAAAAAGUUGAGGAACAAGAAAGCGCACUUCAAAUGCCACUUCAAUGGCGUCAAAUUGU- ((((((((((((((((....))).)))))..(((((.((((.........))))....))))).....)))))))).................((((((......))))))........- ( -32.60) >DroSim_CAF1 99989 119 - 1 GUUCUUCAGAUCUUGCUAUUGCACGGGUUGAUUGGCAGCCGCGAUUAAAACGGCAAGAGUCAAAAAGCUGAAGAACGAGAAGGCGCACUUCAAAUGCCACUUCAAUGGCGUCAAAUUGU- (((((((((...(((((.((((.((..(((((((.......)))))))..)))))).)).)))....)))))))))..((((.....))))..((((((......))))))........- ( -39.70) >DroEre_CAF1 104650 120 - 1 GUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGCUGCGAUUACAACAGCCAGAGUCAAGAAGUUGAAGAACGAGAAAGCGCACUUCAAAUGCCAUUUUAAUGGCGUCAAGUUGUU (((((((((.(((.((....))...(((((.(((.............)))))))).......)))..))))))))).....((((.((((...(((((((....))))))).)))))))) ( -31.82) >DroYak_CAF1 100685 118 - 1 GUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGCCGCGAUUACAACGGCCAGAGUCAA-AAGUUGAAGAACGAGAAAGCGCACUUCAAAUGCCAUUUCAAUGGCGUCAAAUUGU- (((((((((.(((.((..(((..(((((((.....))))).))....)))..)).))).....-...))))))))).................(((((((....)))))))........- ( -31.80) >consensus GUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGCCGCGAUUAAAACGGCAAGAGUCAAAAAGUUGAAGAACGAGAAAGCGCACUUCAAAUGCCAUUUCAAUGGCGUCAAAUUGU_ (((((((((((((.((....))..)))))..(((((.((((.........))))....))))).....))))))))........(((.((...(((((((....)))))))..)).))). (-28.26 = -27.70 + -0.56)

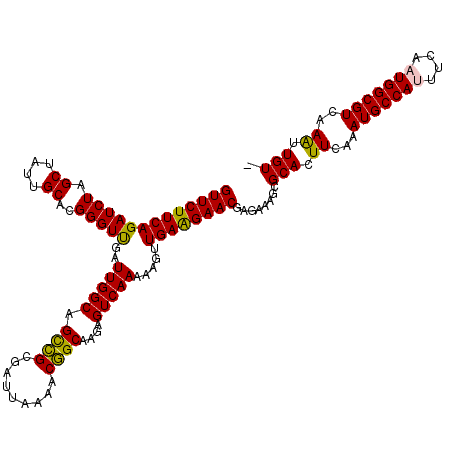
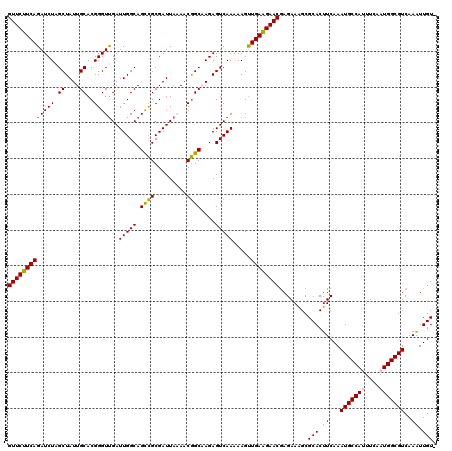
| Location | 716,067 – 716,187 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.29 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 716067 120 + 22407834 UCUCGUUCUUCAACUUUUUGACUCUUGCCGUUUUAAUCGCGACUGCCAAUCAACCCGUGCAAUAGCUAGAUCUGAAGAACUAAAUUCUUUUACUGUUCGUUUUCUCCACGAACUGUCGGA ....((((((((....((((.((.((((((..((.((.((....))..)).))..)).)))).)).))))..)))))))).......((..((.((((((.......)))))).))..)) ( -23.20) >DroSec_CAF1 94841 120 + 1 UCUUGUUCCUCAACUUUUUGACUCUUGCCGUUUUAAUCGCGGCUGCCAAUCGACCCGUGCAAUAGCAAGAUCUGAAGAACUGAAUUCUUUUACUGUUCGUUUUCUCCACGAACUGUCGGA (((((((.....((...((((.....(((((.......)))))......))))...)).....)))))))(((((((((.....))))......((((((.......))))))..))))) ( -24.50) >DroSim_CAF1 100028 120 + 1 UCUCGUUCUUCAGCUUUUUGACUCUUGCCGUUUUAAUCGCGGCUGCCAAUCAACCCGUGCAAUAGCAAGAUCUGAAGAACUGAAUUCUUGUACUGUUCGUUUUCUCCACGAACUGUCGGA ....(((((((((..(((((.((...(((((.......)))))((((.........).)))..))))))).))))))))).......(((.((.((((((.......)))))).))))). ( -30.80) >DroEre_CAF1 104690 120 + 1 UCUCGUUCUUCAACUUCUUGACUCUGGCUGUUGUAAUCGCAGCUGCCAAUCAACCCGUGCAAUAGCUAGAUCUGAAGAACUAAAUUCUUUUACUGUUCGUUUUCUUCACGAACUGUUGGA ....((((((((.......((.(((((((((((((...((....))...........))))))))))))))))))))))).....(((...((.((((((.......)))))).)).))) ( -32.85) >DroYak_CAF1 100724 119 + 1 UCUCGUUCUUCAACUU-UUGACUCUGGCCGUUGUAAUCGCGGCUGCCAAUCAACCCGUGCAAUAGCUAGAUCUGAAGAACUAAAUUCUUUUACUGUUCGUUUUCUUCACGAACUGUUGGA ....((((((((....-..((.((((((..(((((..((.((.((.....)).)))))))))..)))))))))))))))).....(((...((.((((((.......)))))).)).))) ( -32.70) >consensus UCUCGUUCUUCAACUUUUUGACUCUUGCCGUUUUAAUCGCGGCUGCCAAUCAACCCGUGCAAUAGCUAGAUCUGAAGAACUAAAUUCUUUUACUGUUCGUUUUCUCCACGAACUGUCGGA ....((((((((.......((.((((((.(((.....(((((.(........).))))).))).)))))))))))))))).....(((...((.((((((.......)))))).)).))) (-23.81 = -24.29 + 0.48)

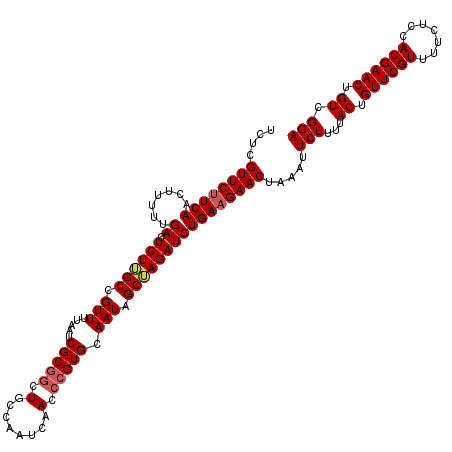
| Location | 716,067 – 716,187 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -29.50 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
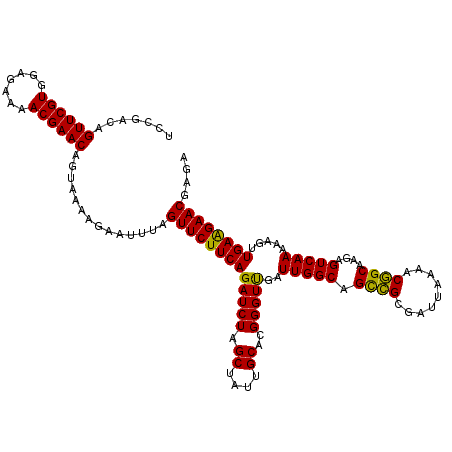
>2L_DroMel_CAF1 716067 120 - 22407834 UCCGACAGUUCGUGGAGAAAACGAACAGUAAAAGAAUUUAGUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGUCGCGAUUAAAACGGCAAGAGUCAAAAAGUUGAAGAACGAGA .......((((((.......))))))..............(((((((((.....(((.((((.((..(((((((.......)))))))..)))))).))).......))))))))).... ( -33.80) >DroSec_CAF1 94841 120 - 1 UCCGACAGUUCGUGGAGAAAACGAACAGUAAAAGAAUUCAGUUCUUCAGAUCUUGCUAUUGCACGGGUCGAUUGGCAGCCGCGAUUAAAACGGCAAGAGUCAAAAAGUUGAGGAACAAGA .......((((((.......))))))..............((((((((((((((((....))).)))))..(((((.((((.........))))....))))).....)))))))).... ( -34.10) >DroSim_CAF1 100028 120 - 1 UCCGACAGUUCGUGGAGAAAACGAACAGUACAAGAAUUCAGUUCUUCAGAUCUUGCUAUUGCACGGGUUGAUUGGCAGCCGCGAUUAAAACGGCAAGAGUCAAAAAGCUGAAGAACGAGA .......((((((.......))))))..........(((.(((((((((...(((((.((((.((..(((((((.......)))))))..)))))).)).)))....)))))))))))). ( -37.40) >DroEre_CAF1 104690 120 - 1 UCCAACAGUUCGUGAAGAAAACGAACAGUAAAAGAAUUUAGUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGCUGCGAUUACAACAGCCAGAGUCAAGAAGUUGAAGAACGAGA .......((((((.......))))))..............(((((((((.(((.((....))...(((((.(((.............)))))))).......)))..))))))))).... ( -29.22) >DroYak_CAF1 100724 119 - 1 UCCAACAGUUCGUGAAGAAAACGAACAGUAAAAGAAUUUAGUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGCCGCGAUUACAACGGCCAGAGUCAA-AAGUUGAAGAACGAGA .......((((((.......))))))..............(((((((((.(((.((..(((..(((((((.....))))).))....)))..)).))).....-...))))))))).... ( -32.20) >consensus UCCGACAGUUCGUGGAGAAAACGAACAGUAAAAGAAUUUAGUUCUUCAGAUCUAGCUAUUGCACGGGUUGAUUGGCAGCCGCGAUUAAAACGGCAAGAGUCAAAAAGUUGAAGAACGAGA .......((((((.......))))))..............(((((((((((((.((....))..)))))..(((((.((((.........))))....))))).....)))))))).... (-29.50 = -28.70 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:04 2006