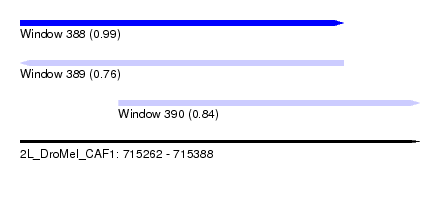
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 715,262 – 715,388 |
| Length | 126 |
| Max. P | 0.988539 |

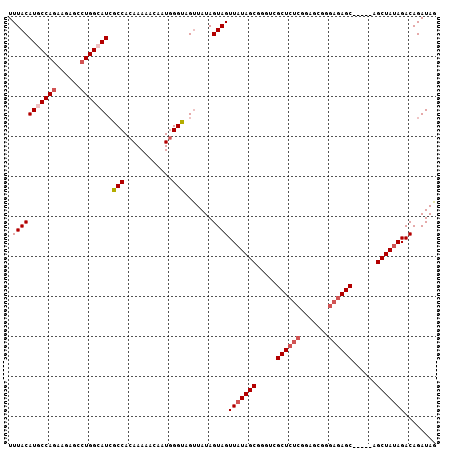
| Location | 715,262 – 715,364 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 715262 102 + 22407834 CUAUCUGUCUAUAGCU-----GCUCUCCCGCUCCGAGAGCGACCCGCUAUAACUACUAUAACUACCUAUUGUUUUUGUGGCGAUGCCAGGCUCUUCUGGCAUGUAAA ...............(-----((((((.......)))))))...((((((((..((.(((......))).))..))))))))((((((((....))))))))..... ( -30.60) >DroSec_CAF1 94006 102 + 1 CUAUCUGUCUAUAGCU-----GCUCUCCCGCUCCGAGAGCGGCCCGCUAUAACUACUAUAGCUACCCAUUGUUUUUGUGGCGAUGCCAGGCUCUUCUGGCAUGUAAA .............(((-----((((((.......))))))))).((((((((..((..............))..))))))))((((((((....))))))))..... ( -35.44) >DroSim_CAF1 99183 102 + 1 CUAUCUGUCUAUAGCU-----GCUCUCCCGCUCCGAGAGCGACCCGCUAUAACUACUAUAACUACCCAUUGUUUUUGUGGCCAUGCCAGGCUCUUCUGGCAUGUAAA .........(((((((-----((((((.......)))))))....)))))).((((...(((........)))...)))).(((((((((....))))))))).... ( -31.90) >DroEre_CAF1 103760 99 + 1 CUAUCUGUCUGUAGCUCUCUCGCU--------CCGAGAGCGGCCCGCUAUAACUACUAUAACUGCCCAUUGUUUUUGUGGCGAUUCCAGGCUCUUGUGGCAUGUAAA .(((.((((....((((((.....--------..))))))((((((((((((..((..............))..))))))))......)))).....)))).))).. ( -26.14) >DroYak_CAF1 99780 107 + 1 CUAUCUGUCUAUAGCUCUCUCGCUCUCUCAGUCCGAGAGCGAACCGCUAUAACUACUAUAACUGCCCAUUGUUUUUGUGGCGAUUCCAGGCUCUUCUGGCAUGUAAA .(((.((((...((((...((((((((.......))))))))..((((((((..((..............))..))))))))......)))).....)))).))).. ( -29.54) >consensus CUAUCUGUCUAUAGCU_____GCUCUCCCGCUCCGAGAGCGACCCGCUAUAACUACUAUAACUACCCAUUGUUUUUGUGGCGAUGCCAGGCUCUUCUGGCAUGUAAA .........((((((......((((((.......)))))).....)))))).((((...(((........)))...))))..((((((((....))))))))..... (-23.82 = -24.70 + 0.88)

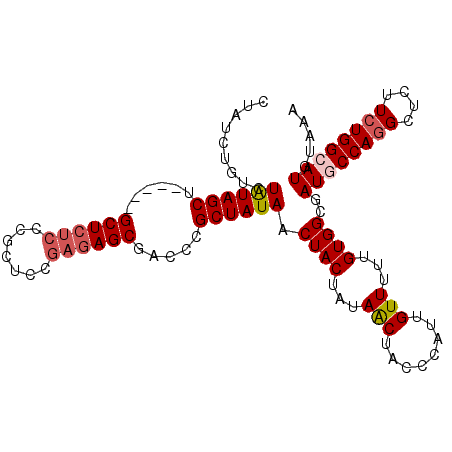
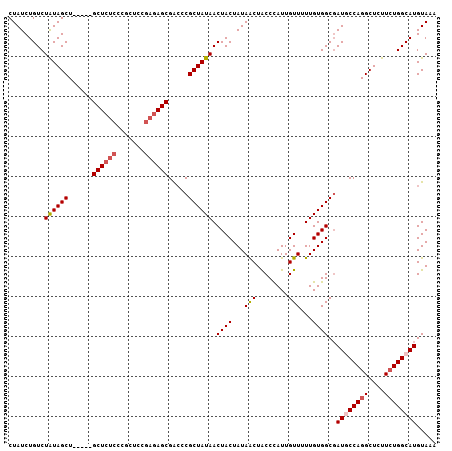
| Location | 715,262 – 715,364 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -24.18 |
| Energy contribution | -25.34 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 715262 102 - 22407834 UUUACAUGCCAGAAGAGCCUGGCAUCGCCACAAAAACAAUAGGUAGUUAUAGUAGUUAUAGCGGGUCGCUCUCGGAGCGGGAGAGC-----AGCUAUAGACAGAUAG .(((((((((((......))))))).(((............))).......))))(((((((.....((((((.......))))))-----.)))))))........ ( -28.40) >DroSec_CAF1 94006 102 - 1 UUUACAUGCCAGAAGAGCCUGGCAUCGCCACAAAAACAAUGGGUAGCUAUAGUAGUUAUAGCGGGCCGCUCUCGGAGCGGGAGAGC-----AGCUAUAGACAGAUAG .....(((((((......))))))).(((.((.......))))).(((((((...))))))).(((.((((((.......))))))-----.)))............ ( -31.70) >DroSim_CAF1 99183 102 - 1 UUUACAUGCCAGAAGAGCCUGGCAUGGCCACAAAAACAAUGGGUAGUUAUAGUAGUUAUAGCGGGUCGCUCUCGGAGCGGGAGAGC-----AGCUAUAGACAGAUAG ....((((((((......))))))))(((.((.......)))))...(((.....(((((((.....((((((.......))))))-----.)))))))....))). ( -32.30) >DroEre_CAF1 103760 99 - 1 UUUACAUGCCACAAGAGCCUGGAAUCGCCACAAAAACAAUGGGCAGUUAUAGUAGUUAUAGCGGGCCGCUCUCGG--------AGCGAGAGAGCUACAGACAGAUAG ..............(.((((......(((.((.......))))).(((((((...))))))))))))((((((..--------.....))))))............. ( -24.10) >DroYak_CAF1 99780 107 - 1 UUUACAUGCCAGAAGAGCCUGGAAUCGCCACAAAAACAAUGGGCAGUUAUAGUAGUUAUAGCGGUUCGCUCUCGGACUGAGAGAGCGAGAGAGCUAUAGACAGAUAG .((((...((((......))))....(((.((.......))))).......))))(((((((..(((((((((.......)))))))))...)))))))........ ( -34.90) >consensus UUUACAUGCCAGAAGAGCCUGGCAUCGCCACAAAAACAAUGGGUAGUUAUAGUAGUUAUAGCGGGUCGCUCUCGGAGCGGGAGAGC_____AGCUAUAGACAGAUAG .(((((((((((......))))))).(((............))).......))))(((((((.....((((((.......))))))......)))))))........ (-24.18 = -25.34 + 1.16)

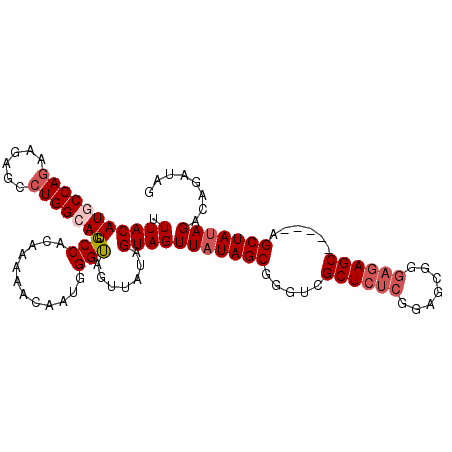
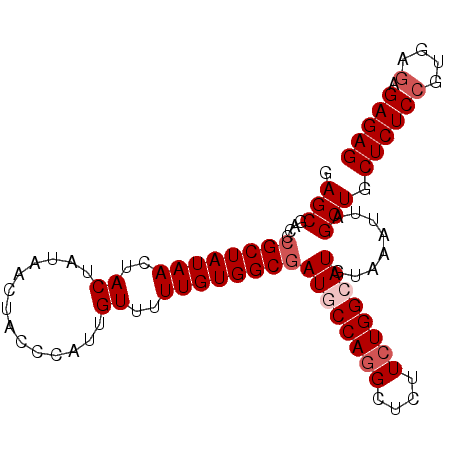
| Location | 715,293 – 715,388 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 715293 95 + 22407834 GAGCGACCCGCUAUAACUACUAUAACUACCUAUUGUUUUUGUGGCGAUGCCAGGCUCUUCUGGCAUGUAAAUUAGCUGCUCUCCGUGAGAGAGAG .(((....((((((((..((.(((......))).))..))))))))((((((((....))))))))........))).((((((....).))))) ( -33.10) >DroSec_CAF1 94037 95 + 1 GAGCGGCCCGCUAUAACUACUAUAGCUACCCAUUGUUUUUGUGGCGAUGCCAGGCUCUUCUGGCAUGUAAAUUAGCUGCUCUCCGUGAGAGAGAG ..(((((.((((((((..((..............))..))))))))((((((((....))))))))........)))))(((((....).)))). ( -34.64) >DroSim_CAF1 99214 95 + 1 GAGCGACCCGCUAUAACUACUAUAACUACCCAUUGUUUUUGUGGCCAUGCCAGGCUCUUCUGGCAUGUAAAUUAGCUGCUCUCCGUGAGAGAGAG .(((.....(((((((..((..............))..)))))))(((((((((....))))))))).......))).((((((....).))))) ( -31.54) >DroEre_CAF1 103788 95 + 1 GAGCGGCCCGCUAUAACUACUAUAACUGCCCAUUGUUUUUGUGGCGAUUCCAGGCUCUUGUGGCAUGUAAAUUAGCUGCUCUCGGCGAGAGAGAG (((((((.((((((((..((..............))..))))))))....((.(((.....))).)).......)))))))((..(....).)). ( -24.54) >DroYak_CAF1 99816 95 + 1 GAGCGAACCGCUAUAACUACUAUAACUGCCCAUUGUUUUUGUGGCGAUUCCAGGCUCUUCUGGCAUGUAAAUUAGCUGCUCUCCGCGAGAGAGAG (((((((.((((((((..((..............))..)))))))).)))...))))....(((..........))).((((((....).))))) ( -24.74) >consensus GAGCGACCCGCUAUAACUACUAUAACUACCCAUUGUUUUUGUGGCGAUGCCAGGCUCUUCUGGCAUGUAAAUUAGCUGCUCUCCGUGAGAGAGAG .(((....((((((((..((..............))..))))))))((((((((....))))))))........))).(((((.....))))).. (-24.88 = -25.68 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:01 2006