
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,614,070 – 6,614,205 |
| Length | 135 |
| Max. P | 0.956029 |

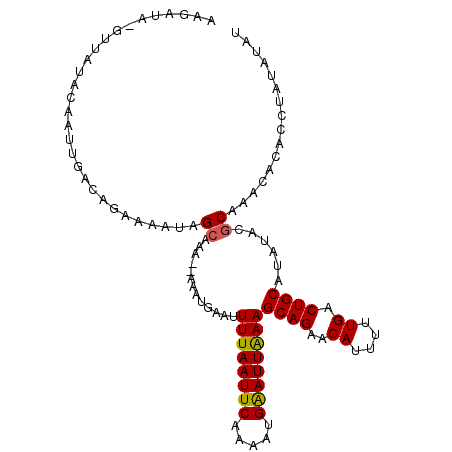
| Location | 6,614,070 – 6,614,176 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -13.82 |
| Consensus MFE | -11.16 |
| Energy contribution | -10.97 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
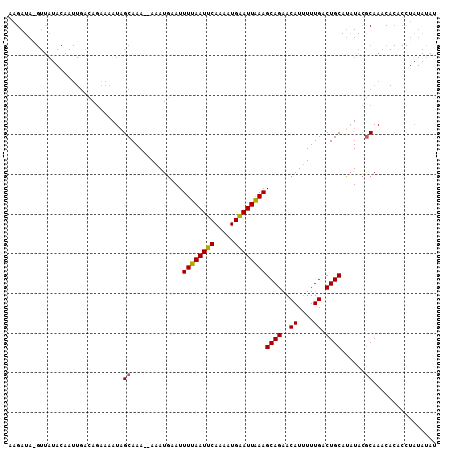
>2L_DroMel_CAF1 6614070 106 - 22407834 AAGAUAGGUUAUACAAUUGACAGAAAAUAGCAAA--AAAUGAAUUUUAAUUCAAAAUGAAUUAAAGCAGAACAUUUUUGACUGCAUAUACGCAAAUACACCUAUACAU ...(((((((((...(((.......))).((...--........((((((((.....))))))))((((..((....)).))))......))..))).)))))).... ( -16.50) >DroSec_CAF1 41190 82 - 1 AAGAUA------------GACAGAAAAUAGCAAA--AAAUGAAUUUUAAUUCAAAAUGAAUUAAAGCAGAACAUUUUUGACUGCAUAUACGCAAAC------------ ......------------...........((...--........((((((((.....))))))))((((..((....)).))))......))....------------ ( -12.50) >DroSim_CAF1 42401 108 - 1 AAGAUACGUUAUACAAUUUACAGAAAAUAGCAAAAAAAAUGAAUUUUAAUUCAAAAUGGAUUGAAGCAGAACAUUUUUGACUGCAUAUACGCAAGCACACCUAUAUAU .............................(((..(((((((..(((((((((.....))))))))).....)))))))...))).....(....)............. ( -12.50) >DroYak_CAF1 43016 107 - 1 AAGAUAAGUUAUACAAUUGAGAUAAAAUUGCAAAA-AAAUUAAUUUUAAUUCAAACUGAAUUGAAGCAGCACAUAUUUGACUGCAGCUAUACAAACACGCCUAUAUAU ......((((...(((((.......))))).....-........((((((((.....))))))))((((((......)).)))))))).................... ( -13.80) >consensus AAGAUA_GUUAUACAAUUGACAGAAAAUAGCAAA__AAAUGAAUUUUAAUUCAAAAUGAAUUAAAGCAGAACAUUUUUGACUGCAUAUACGCAAACACACCUAUAUAU .............................((.............((((((((.....))))))))((((..((....)).))))......))................ (-11.16 = -10.97 + -0.19)

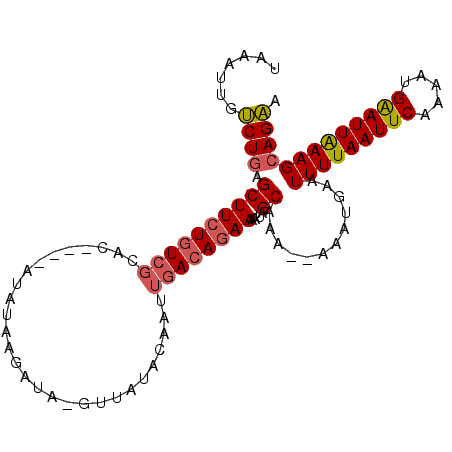
| Location | 6,614,107 – 6,614,205 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -12.38 |
| Energy contribution | -12.81 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
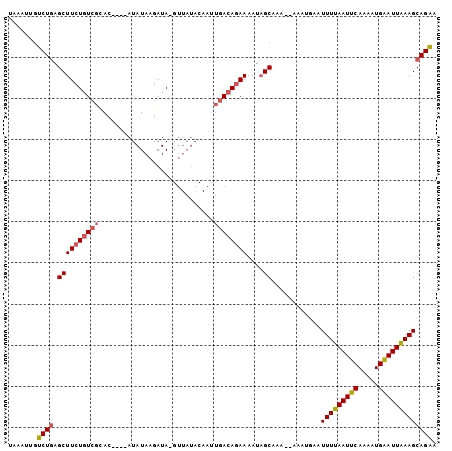
>2L_DroMel_CAF1 6614107 98 - 22407834 CGAUUUGUCUGAGCUUCUGUCGCAC----AUAUAAGAUAGGUUAUACAAUUGACAGAAAAUAGCAAA--AAAUGAAUUUUAAUUCAAAAUGAAUUAAAGCAGAA .......((((.((((((((((...----.(((((......)))))....)))))))....)))...--.......(((((((((.....))))))))))))). ( -20.40) >DroSec_CAF1 41215 86 - 1 UAAAUUGUCUGAGCUUCUGUCGCAC----AUAUAAGAUA------------GACAGAAAAUAGCAAA--AAAUGAAUUUUAAUUCAAAAUGAAUUAAAGCAGAA .......((((.(((((((((.(..----......)...------------))))))....)))...--.......(((((((((.....))))))))))))). ( -18.70) >DroSim_CAF1 42438 100 - 1 UAAAUUGUCUGAGCUUCUGUCGCAC----AUAUAAGAUACGUUAUACAAUUUACAGAAAAUAGCAAAAAAAAUGAAUUUUAAUUCAAAAUGGAUUGAAGCAGAA .......((((.((((((((.....----.(((((......)))))......)))))....)))............(((((((((.....))))))))))))). ( -15.80) >DroYak_CAF1 43053 103 - 1 GGAUUUAGCUAAGCUUCUGUCGUACAUACAUAUAAGAUAAGUUAUACAAUUGAGAUAAAAUUGCAAAA-AAAUUAAUUUUAAUUCAAACUGAAUUGAAGCAGCA .......(((..(((((..(((((...((.(((...))).))..)))..(((((.((((((((.....-....)))))))).)))))...))...)))))))). ( -16.70) >consensus UAAAUUGUCUGAGCUUCUGUCGCAC____AUAUAAGAUA_GUUAUACAAUUGACAGAAAAUAGCAAA__AAAUGAAUUUUAAUUCAAAAUGAAUUAAAGCAGAA .......((((.((((((((((............................))))))))....))............(((((((((.....))))))))))))). (-12.38 = -12.81 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:46 2006