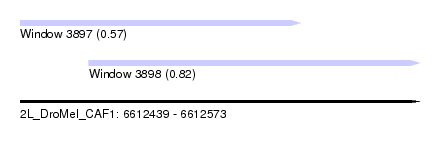
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,612,439 – 6,612,573 |
| Length | 134 |
| Max. P | 0.824539 |

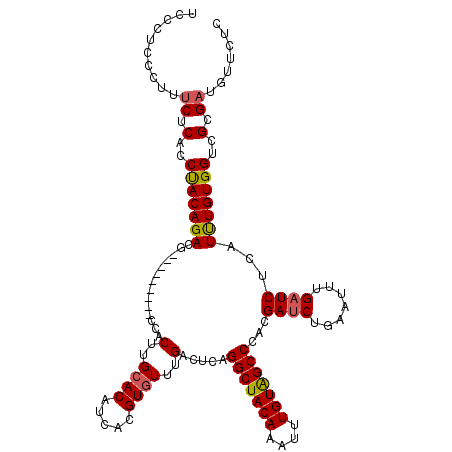
| Location | 6,612,439 – 6,612,533 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -12.57 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6612439 94 + 22407834 AACAACACUUACGAUUAUGUAUUUCCCUCCCUUUCUCACCUACAGACG---------CCACUUGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUGGCCC ..........................................((((((---------(..............))))))).....(((((((....))))))). ( -15.54) >DroSec_CAF1 39724 94 + 1 AACAACACUUACGACUAUGUAUUUCUCUCCCUUUCUCACCUACAGACG---------CCACUUGCACAUCACGUUUUUGACUCAGGCUACAAAUUUGUAGCCC .....((...(((...((((.............(((.......))).(---------(.....))))))..)))...)).....(((((((....))))))). ( -13.50) >DroSim_CAF1 40906 94 + 1 AACAACACUUACGACUAUGUAUUUCCGUCCCUUUCUCACCUACAGACG---------CCACUUGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUAGCCC .....((..((((....((((....((((...............))))---------.....)))).....))))..)).....(((((((....))))))). ( -18.16) >DroEre_CAF1 40790 103 + 1 AGCAACACUUACGAUUAUGUAUUUCCCUCUCUUCCUCGCCCACAGACAAGGCGGUCGGCACUCGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUGGCCC .....((..((((...((((...............(((((.........)))))...((....))))))..))))..)).....(((((((....))))))). ( -22.40) >consensus AACAACACUUACGACUAUGUAUUUCCCUCCCUUUCUCACCUACAGACG_________CCACUUGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUAGCCC ............................................................(..((((.....))))..).....(((((((....))))))). (-12.57 = -12.57 + 0.00)

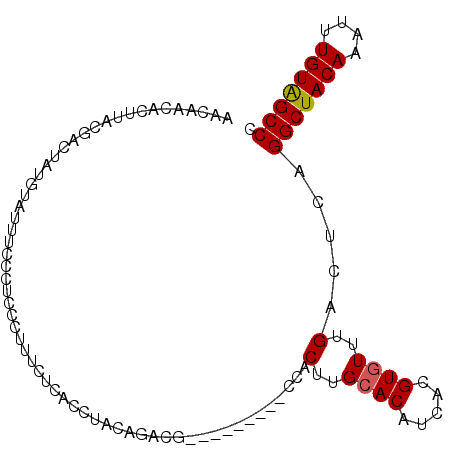
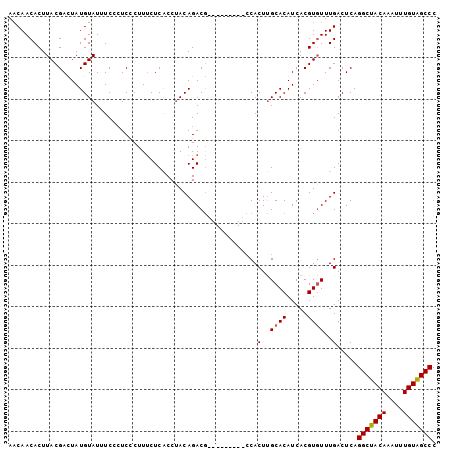
| Location | 6,612,462 – 6,612,573 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.30 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -22.15 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6612462 111 + 22407834 UCCCUCCCUUUCUCACCUACAGACG---------CCACUUGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUGGCCCACGAUCUGAAUUUGAUCUCAUCUGUGGUCGCGAUGUUCUC ..........((.(..(((((((..---------...(..((((.....))))..).....(((((((....)))))))...((((.......))))...)))))))..).))....... ( -26.70) >DroSec_CAF1 39747 111 + 1 UCUCUCCCUUUCUCACCUACAGACG---------CCACUUGCACAUCACGUUUUUGACUCAGGCUACAAAUUUGUAGCCCACGAUCUGAAUUUGAUCUCAUUUGUGGUCGCGAUGUUCUC ..................(((..((---------((((.((...((((.((((..((.((.(((((((....)))))))...)))).)))).))))..))...)))).))...))).... ( -22.90) >DroSim_CAF1 40929 111 + 1 UCCGUCCCUUUCUCACCUACAGACG---------CCACUUGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUAGCCCACGAUCUGAAUUUGAUCUCAUUUGUGGUCGCGAUGUUCUC ..((((.............((((((---------...(..((((.....))))..).....(((((((....)))))))..)).)))).....((((.(....).))))..))))..... ( -25.50) >DroEre_CAF1 40813 114 + 1 UCCCUCUCUUCCUCGCCCACAGACAAGGCGGUCGGCACUCGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUGGCCCACGAUCU------GCUCUCAUUUGUGGUUGCGAUGUUCUC ............(((((((((((..(((((((((...(..((((.....))))..).....(((((((....)))))))..))).))------)).))..)))))))..))))....... ( -36.90) >consensus UCCCUCCCUUUCUCACCUACAGACG_________CCACUUGCACAUCACGUGUUUGACUCAGGCUACAAAUUUGUAGCCCACGAUCUGAAUUUGAUCUCAUUUGUGGUCGCGAUGUUCUC ..........((.(..(((((((..............(..((((.....))))..).....(((((((....)))))))...((((.......))))...)))))))..).))....... (-22.15 = -22.28 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:40 2006