
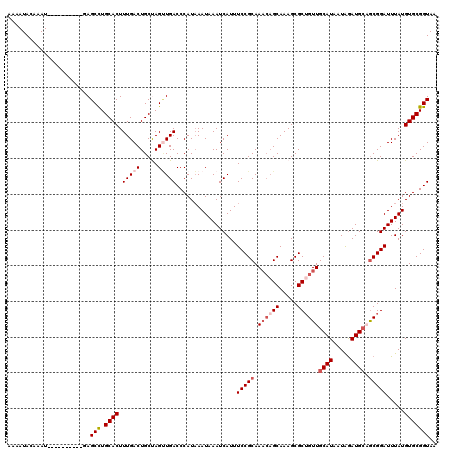
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,610,155 – 6,610,302 |
| Length | 147 |
| Max. P | 0.988407 |

| Location | 6,610,155 – 6,610,262 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.86 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6610155 107 + 22407834 AAAAUACAAAU----------AAGCCUGCACUUUGACUGCUAGUUGACCCAUAAAUAAAUCAUUUCCGCAA---GCAAAGCGCUGUUGCAUAAUAGAUGCAACGGAUUUAUGUGCGGUAA ...........----------..(((.((((.(..((.....))..).......(((((((.....(((..---.....))).((((((((.....)))))))))))))))))))))).. ( -26.60) >DroSec_CAF1 37383 110 + 1 AAAAUACAAAU----------GAGCCUGCACUUUGACUGCCAGUUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCAGUUGCAUAAUAGAUGCAGCGGAUUUAUGUGCGGUAA ...........----------..(((.((((.(..((.....))..).......(((((((.....(((..........)))..(((((((.....))))))).)))))))))))))).. ( -26.10) >DroSim_CAF1 38607 110 + 1 AAAAUACAAAU----------GAGCCUGCACUUUGACUGCCAGUUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUAGAUGCAGCGGAUUUAUGUGCGGUAA ...........----------..(((.((((.(..((.....))..).................(((((.((((((.....))))))((((.....)))).))))).....))))))).. ( -30.10) >DroEre_CAF1 38567 119 + 1 AAAAUACAAAUGAGCUGAGUUGAGCUUGCACUUUGACUGCUAGCUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUAGAUG-UGCGGAUUUAUGUGCGGUAA .......((((((..((.(((.((((.(((.......))).))))))).))........))))))(((((((((((.....))))))(((((.....))-))).........)))))... ( -29.50) >DroYak_CAF1 39133 120 + 1 AAAAUACAAAUUUGCUGAGUUGAGCCUGCACUUUGACUGCUAGGUGACCCAUUAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUGAAUGCCGCGGAUUUAUGUGCGGUAA ...........(((.((.(((..(((((((.......))).))))))).)).)))....(((((...((((.((((.....))))))))..))))).(((((((.(....).))))))). ( -30.80) >consensus AAAAUACAAAU__________GAGCCUGCACUUUGACUGCUAGUUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUAGAUGCAGCGGAUUUAUGUGCGGUAA .......................(((.((((.(((((.....))))).................(((((.((((((.....))))))((((.....)))).))))).....))))))).. (-21.42 = -22.86 + 1.44)


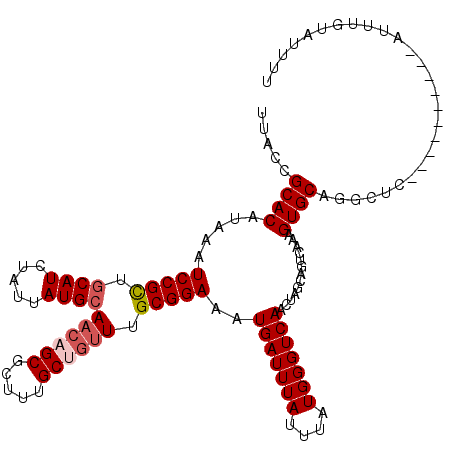
| Location | 6,610,155 – 6,610,262 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6610155 107 - 22407834 UUACCGCACAUAAAUCCGUUGCAUCUAUUAUGCAACAGCGCUUUGC---UUGCGGAAAUGAUUUAUUUAUGGGUCAACUAGCAGUCAAAGUGCAGGCUU----------AUUUGUAUUUU .....(((.((((..(((((((((.....))))))).((((((((.---((((.....(((((((....)))))))....)))).)))))))).)).))----------)).)))..... ( -32.80) >DroSec_CAF1 37383 110 - 1 UUACCGCACAUAAAUCCGCUGCAUCUAUUAUGCAACUGCGCUUUGCUGUUUGCGGAAAUGAUUUAUUUAUGGGUCAACUGGCAGUCAAAGUGCAGGCUC----------AUUUGUAUUUU .........((((((..(((((((.....))))).((((((((((((((..(..((.((((.....))))...))..)..)))).))))))))))))..----------))))))..... ( -32.70) >DroSim_CAF1 38607 110 - 1 UUACCGCACAUAAAUCCGCUGCAUCUAUUAUGCAACAGCGCUUUGCUGUUUGCGGAAAUGAUUUAUUUAUGGGUCAACUGGCAGUCAAAGUGCAGGCUC----------AUUUGUAUUUU .........((((((..(((((((.....))))....((((((((((((..(..((.((((.....))))...))..)..)))).)))))))).)))..----------))))))..... ( -29.20) >DroEre_CAF1 38567 119 - 1 UUACCGCACAUAAAUCCGCA-CAUCUAUUAUGCAACAGCGCUUUGCUGUUUGCGGAAAUGAUUUAUUUAUGGGUCAGCUAGCAGUCAAAGUGCAAGCUCAACUCAGCUCAUUUGUAUUUU .....((..........)).-........((((((..(((((((((((((.((.((.((((.....))))...)).)).))))).)))))))).((((......))))...))))))... ( -29.00) >DroYak_CAF1 39133 120 - 1 UUACCGCACAUAAAUCCGCGGCAUUCAUUAUGCAACAGCGCUUUGCUGUUUGCGGAAAUGAUUUAUUAAUGGGUCACCUAGCAGUCAAAGUGCAGGCUCAACUCAGCAAAUUUGUAUUUU ...((((..........))))........((((((..(((((((((((((...((...(((((((....))))))))).))))).))))))))..(((......)))....))))))... ( -31.50) >consensus UUACCGCACAUAAAUCCGCUGCAUCUAUUAUGCAACAGCGCUUUGCUGUUUGCGGAAAUGAUUUAUUUAUGGGUCAACUAGCAGUCAAAGUGCAGGCUC__________AUUUGUAUUUU .....((((.....(((((.((((.....))))((((((.....)))))).)))))..(((((((....))))))).............))))........................... (-23.28 = -24.12 + 0.84)

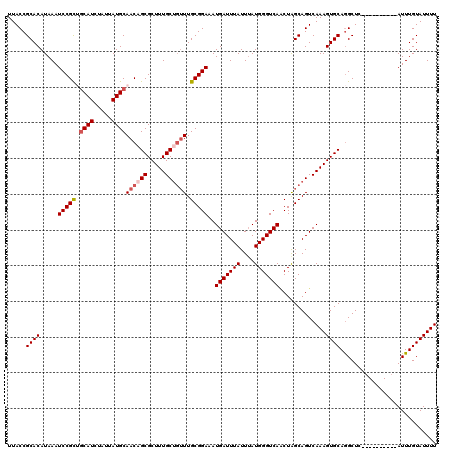
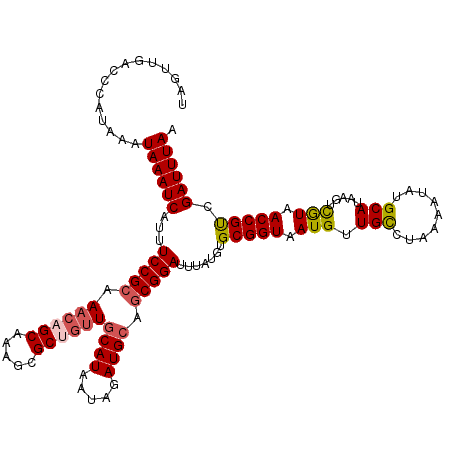
| Location | 6,610,185 – 6,610,302 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -25.74 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6610185 117 + 22407834 UAGUUGACCCAUAAAUAAAUCAUUUCCGCAA---GCAAAGCGCUGUUGCAUAAUAGAUGCAACGGAUUUAUGUGCGGUAAUGUUGUCUAAAAUAUGCAUAAGUCGUAACCGUCGAUUUAA .(((((((............((((.(((((.---..(((...(((((((((.....))))))))).)))...))))).))))..........(((((....).))))...)))))))... ( -29.30) >DroSec_CAF1 37413 120 + 1 CAGUUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCAGUUGCAUAAUAGAUGCAGCGGAUUUAUGUGCGGUAAUGUUGCCUAAAAAAUGCAUAAGUCGUAACCGUCGAUUUAA ...............((((((...(((((.....((.....))...(((((.....)))))))))).......(((((.(((.(((.........))).....))).))))).)))))). ( -27.00) >DroSim_CAF1 38637 120 + 1 CAGUUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUAGAUGCAGCGGAUUUAUGUGCGGUAAUGUUGCCUAAAAUAUGCAUAAGUCGUAACCGUCGAUUUAA ...............((((((...(((((.((((((.....))))))((((.....)))).))))).......(((((.(((.(((.........))).....))).))))).)))))). ( -32.00) >DroEre_CAF1 38607 119 + 1 UAGCUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUAGAUG-UGCGGAUUUAUGUGCGGUAAUGUUGCCUAAAAUAUGCAUAAGUUAUAACCGUCGAUUUAA ...............((((((...(((((.((((((.....))))))((((.....)))-)))))).......(((((.(((.(((.........))).....))).))))).)))))). ( -28.20) >DroYak_CAF1 39173 120 + 1 UAGGUGACCCAUUAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUGAAUGCCGCGGAUUUAUGUGCGGUAAUGUUGUCUAAAAUAUGCAUAAGUUGUAACCGCCGAUUUAA ..(....).......((((((...(((((.((((((.....))))))((((.....)))).))))).....(.(((((..((((......))))((((.....)))))))))))))))). ( -33.80) >consensus UAGUUGACCCAUAAAUAAAUCAUUUCCGCAAACAGCAAAGCGCUGUUGCAUAAUAGAUGCAGCGGAUUUAUGUGCGGUAAUGUUGCCUAAAAUAUGCAUAAGUCGUAACCGUCGAUUUAA ...............((((((...(((((.((((((.....))))))((((.....)))).))))).......(((((.(((.(((.........))).....))).))))).)))))). (-25.74 = -26.14 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:39 2006