
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
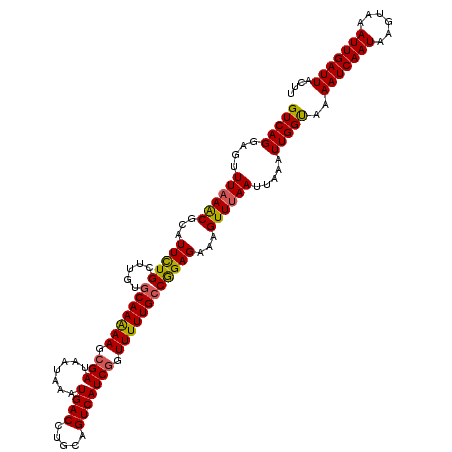
| Location | 6,608,513 – 6,608,633 |
| Length | 120 |
| Max. P | 0.846221 |

| Location | 6,608,513 – 6,608,633 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -22.23 |
| Energy contribution | -21.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6608513 120 + 22407834 GUCAGGAGUUUUAACGCAUUCUGCUUGUACAAAAAGCGAUAAUAAAUGACCUGCAGUCAUCGGUUUUUGCCGGAGAAAGUUUAAUUAAAUUGGUAAAAUCAAUAAGUAAAUUGAUUACUU (((((((((........)))))((((.......)))).........))))..((..((..((((....))))..))..))...........((((..((((((......)))))))))). ( -20.70) >DroSec_CAF1 35727 120 + 1 GUCAGCAGUUUAAACGCAUUCUGCUUGUGCAAAAAGCGAUAAUAAAUGACCUGCAGUCAUCGGUUUUUGCCGGAGAAAGUUUAAUUAAAUUGGUAAAAUCAAUAAGUAAAUUGAUUACUU .....((((((((..((.((((.((.(.(((((((.(((.......((((.....))))))).)))))))))))))).))....))))))))....(((((((......))))))).... ( -25.31) >DroSim_CAF1 36920 120 + 1 GUCAGCAGUUUAAACGCAUUCUGCUUGUGCAAAAAGCGAUAAUAAAUGACCUGCAGUCAUCGGUUUUUGCCGGAGAAAGUUUAAUUAAAUUGGUAAAAUCAAUAAGUAAAUUGAUUACUU .....((((((((..((.((((.((.(.(((((((.(((.......((((.....))))))).)))))))))))))).))....))))))))....(((((((......))))))).... ( -25.31) >DroEre_CAF1 36862 120 + 1 GUCAGGAGUUUAAGCGCAUUCUGCUUGUGCAAAAAGCGAUAAUAAAUGACCUGCAGUCAUCGCUUUUUGCCAGAGAAAGUUUAAUUAAAUUGGCAAAAUCAAUAAGUAAAUUGAUUACUU (((((....((((((...((((.((.(.(((((((((((.......((((.....)))))))))))))))))))))).)))))).....)))))..(((((((......))))))).... ( -33.71) >DroYak_CAF1 37527 120 + 1 GUCAGGAGCUUAAACGCAUUUUGCUUGUGCAAGAAGCGAUAAUAAAUGACCUGCAGUCAUCACUUUUUGCCAGAGAAAGUUUAAUUAAAUUGGCAAAAUCAAUAGGUAAAUUGAUUACUU (((((..(((((..(((.((((((....)))))).))).......(((((.....)))))....(((((((((...((......))...)))))))))....)))))...)))))..... ( -27.40) >consensus GUCAGGAGUUUAAACGCAUUCUGCUUGUGCAAAAAGCGAUAAUAAAUGACCUGCAGUCAUCGGUUUUUGCCGGAGAAAGUUUAAUUAAAUUGGUAAAAUCAAUAAGUAAAUUGAUUACUU (((((....((((((...(((((.....(((((((.(((.......((((.....))))))).))))))))))))...)))))).....)))))..(((((((......))))))).... (-22.23 = -21.87 + -0.36)

| Location | 6,608,513 – 6,608,633 |
|---|---|
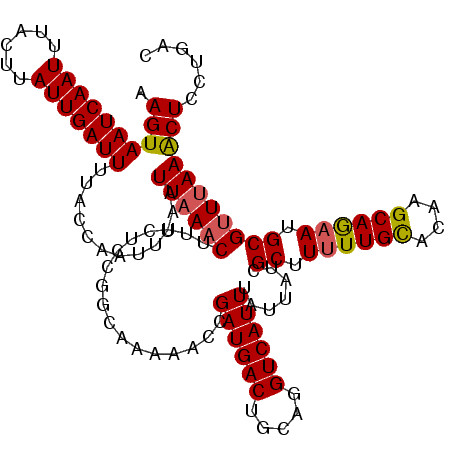
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6608513 120 - 22407834 AAGUAAUCAAUUUACUUAUUGAUUUUACCAAUUUAAUUAAACUUUCUCCGGCAAAAACCGAUGACUGCAGGUCAUUUAUUAUCGCUUUUUGUACAAGCAGAAUGCGUUAAAACUCCUGAC ....(((((((......)))))))....................((..(((......)))..))...((((...((((....(((.((((((....)))))).))).))))...)))).. ( -21.70) >DroSec_CAF1 35727 120 - 1 AAGUAAUCAAUUUACUUAUUGAUUUUACCAAUUUAAUUAAACUUUCUCCGGCAAAAACCGAUGACUGCAGGUCAUUUAUUAUCGCUUUUUGCACAAGCAGAAUGCGUUUAAACUGCUGAC ....(((((((......)))))))........................(((((......((((((.....))))))..(((.(((.((((((....)))))).)))..)))..))))).. ( -24.80) >DroSim_CAF1 36920 120 - 1 AAGUAAUCAAUUUACUUAUUGAUUUUACCAAUUUAAUUAAACUUUCUCCGGCAAAAACCGAUGACUGCAGGUCAUUUAUUAUCGCUUUUUGCACAAGCAGAAUGCGUUUAAACUGCUGAC ....(((((((......)))))))........................(((((......((((((.....))))))..(((.(((.((((((....)))))).)))..)))..))))).. ( -24.80) >DroEre_CAF1 36862 120 - 1 AAGUAAUCAAUUUACUUAUUGAUUUUGCCAAUUUAAUUAAACUUUCUCUGGCAAAAAGCGAUGACUGCAGGUCAUUUAUUAUCGCUUUUUGCACAAGCAGAAUGCGCUUAAACUCCUGAC ((((((.....))))))......(((((((..................)))))))((((((((((.....)))))).......((.((((((....)))))).))))))........... ( -27.67) >DroYak_CAF1 37527 120 - 1 AAGUAAUCAAUUUACCUAUUGAUUUUGCCAAUUUAAUUAAACUUUCUCUGGCAAAAAGUGAUGACUGCAGGUCAUUUAUUAUCGCUUCUUGCACAAGCAAAAUGCGUUUAAGCUCCUGAC .(((..(((((......)))))((((((((..................)))))))).......))).((((...((((....(((...((((....))))...)))..))))..)))).. ( -23.77) >consensus AAGUAAUCAAUUUACUUAUUGAUUUUACCAAUUUAAUUAAACUUUCUCCGGCAAAAACCGAUGACUGCAGGUCAUUUAUUAUCGCUUUUUGCACAAGCAGAAUGCGUUUAAACUCCUGAC .((((((((((......)))))))............((((((.................((((((.....)))))).......((.((((((....)))))).)))))))))))...... (-19.54 = -19.66 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:36 2006