
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,589,662 – 6,589,754 |
| Length | 92 |
| Max. P | 0.746587 |

| Location | 6,589,662 – 6,589,754 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
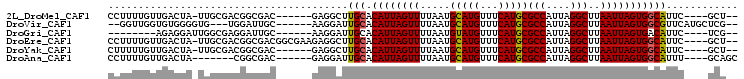
>2L_DroMel_CAF1 6589662 92 + 22407834 CCUUUUGUUGACUA-UUGCGACGGCGAC------GAGGCUUGCACAUUAGUUUUAAUGCAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGGCAUUC----GCU-- ......((((.(..-..)))))(((((.------(((((.((((............)))).)))))....((((((((......))))))))..))----)))-- ( -26.40) >DroVir_CAF1 21957 92 + 1 --GGUUGGUGUGGGGUG---UGGAUUGC------AAGGAUUGCACAUUAGUUUUAAUGCAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGGCGUUCAUGCUCG-- --..........(((((---(((((.((------(.(((.((((............))))...))).)))((((((((......)))))))))))))))))).-- ( -27.90) >DroGri_CAF1 25169 85 + 1 --------AGAGGAUUGGCGAGGAUUGC------AAGGAUUGCACAUUAGUUUUAAUGUAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGACAUUC----UCG-- --------..........((((((((((------(.....))))((((((((.....(((((...)))))(((....)))..))))))))..))))----)))-- ( -20.70) >DroEre_CAF1 17917 98 + 1 CCUUUUGUUGACUA-UUGCGACGGCGACGGCGAAGAGGCUUGCACAUUAGUUUUAAUGCAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGGCAUUC----GCU-- ....((((((.(..-..)))))))....(((((((((((.((((............)))).)))))....((((((((......)))))))).)))----)))-- ( -28.00) >DroYak_CAF1 17799 92 + 1 CUUUUUGUUGACUA-UUGCGACGGCGAC------GAGGCUUGCACAUUAGUUUUAAUGCAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGGCAUUC----GCU-- ......((((.(..-..)))))(((((.------(((((.((((............)))).)))))....((((((((......))))))))..))----)))-- ( -26.40) >DroAna_CAF1 18387 88 + 1 CCUUUUGUUGACUA-------CGGCGAC------GAGGAUUGCACAUUAGUUUUAAUGCAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGGCAUUU----GCAGC ((((..((((.(..-------..)))))------)))).(((((.............(((((...)))))((((((((......))))))))...)----)))). ( -26.70) >consensus CCUUUUGUUGACUA_UUGCGACGGCGAC______GAGGAUUGCACAUUAGUUUUAAUGCAUGUUUCAUGCGCCAUUAGGCUUAAUUAGUGGCAUUC____GCU__ ........................................(((.((((((((.....(((((...)))))(((....)))..)))))))))))............ (-16.14 = -16.03 + -0.11)

| Location | 6,589,662 – 6,589,754 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.75 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
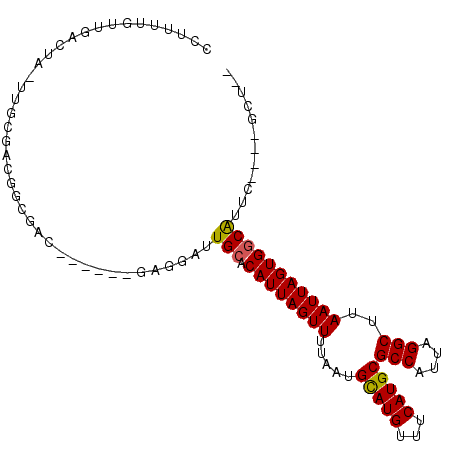
>2L_DroMel_CAF1 6589662 92 - 22407834 --AGC----GAAUGCCACUAAUUAAGCCUAAUGGCGCAUGAAACAUGCAUUAAAACUAAUGUGCAAGCCUC------GUCGCCGUCGCAA-UAGUCAACAAAAGG --.((----((..((..........))....(((((.((((..(.(((((..........))))).)..))------)))))))))))..-.............. ( -19.90) >DroVir_CAF1 21957 92 - 1 --CGAGCAUGAACGCCACUAAUUAAGCCUAAUGGCGCAUGAAACAUGCAUUAAAACUAAUGUGCAAUCCUU------GCAAUCCA---CACCCCACACCAACC-- --...(((.((..((((........(((....)))(((((...)))))...........)).))..))..)------))......---...............-- ( -16.00) >DroGri_CAF1 25169 85 - 1 --CGA----GAAUGUCACUAAUUAAGCCUAAUGGCGCAUGAAACAUACAUUAAAACUAAUGUGCAAUCCUU------GCAAUCCUCGCCAAUCCUCU-------- --(((----(...............(((....)))(((.......(((((((....))))))).......)------))....))))..........-------- ( -16.54) >DroEre_CAF1 17917 98 - 1 --AGC----GAAUGCCACUAAUUAAGCCUAAUGGCGCAUGAAACAUGCAUUAAAACUAAUGUGCAAGCCUCUUCGCCGUCGCCGUCGCAA-UAGUCAACAAAAGG --.((----((..((.((..((((....))))((((...((..(.(((((..........))))).)..))..)))))).))..))))..-.............. ( -20.50) >DroYak_CAF1 17799 92 - 1 --AGC----GAAUGCCACUAAUUAAGCCUAAUGGCGCAUGAAACAUGCAUUAAAACUAAUGUGCAAGCCUC------GUCGCCGUCGCAA-UAGUCAACAAAAAG --.((----((..((..........))....(((((.((((..(.(((((..........))))).)..))------)))))))))))..-.............. ( -19.90) >DroAna_CAF1 18387 88 - 1 GCUGC----AAAUGCCACUAAUUAAGCCUAAUGGCGCAUGAAACAUGCAUUAAAACUAAUGUGCAAUCCUC------GUCGCCG-------UAGUCAACAAAAGG (((((----....((..........)).....((((.((((....(((((..........)))))....))------)))))))-------)))).......... ( -18.00) >consensus __AGC____GAAUGCCACUAAUUAAGCCUAAUGGCGCAUGAAACAUGCAUUAAAACUAAUGUGCAAGCCUC______GUCGCCGUCGCAA_UAGUCAACAAAAGG ............((.(((.......(((....)))(((((...)))))............)))))........................................ (-11.42 = -11.75 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:28 2006