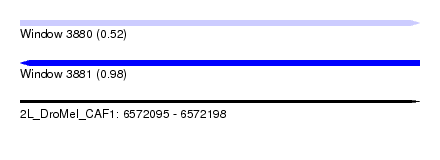
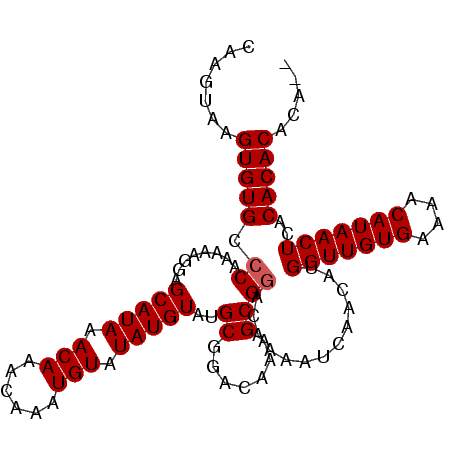
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,572,095 – 6,572,198 |
| Length | 103 |
| Max. P | 0.984726 |

| Location | 6,572,095 – 6,572,198 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6572095 103 + 22407834 --UUUGUGUGUUAGUUAUGUUUUCACAACCAUGUUGAUUUUGUCCGCUUUGUCCGCAUACAUAUACAUUUGUUUGUUUAUGCUCCUUUUUGGGCACACUUACUUG --...((((((......(((....))).((((((.(((...(....)...))).)))).((((.(((......))).)))).........))))))))....... ( -17.40) >DroSim_CAF1 46135 103 + 1 --UGUGUGUGUGAGUUAUGUUUUCACAACCAUGUUGAUUUUGUCCGCUUUGUCCGCAUACAUAUACAUUUGUUUGUUUAUGCUCCUUUUUGGGCACACUUACUUG --.(((((((((((.......)))))..((((((.(((...(....)...))).)))).((((.(((......))).)))).........))))))))....... ( -22.80) >DroEre_CAF1 45013 105 + 1 CGUGUGUGUGUGAGUUAUGUUUUCACAACCAUGUUGAUUUUGUCCGCUUUGUCCGCAUACAUAUACAUUUGUUUGUUUAUGCUCCUUUUUGGGCACACUUACUUG .((((((.((((((.......)))))).((((((.(((...(....)...))).)))).((((.(((......))).)))).........))))))))....... ( -24.00) >DroAna_CAF1 46963 103 + 1 --UGUGUGUGUGAGUUAUGUUUUCACAACCAUGUUGAUUUUGUUCGCUUUGUCCGCAUACAUAUACAGUUAUUUGUUUAUGCUUCUCUUUGUGCACACUUACUUG --.(((((((((((.......)))))....((((.(((...(....)...))).)))).((((.((((....)))).))))...........))))))....... ( -20.40) >consensus __UGUGUGUGUGAGUUAUGUUUUCACAACCAUGUUGAUUUUGUCCGCUUUGUCCGCAUACAUAUACAUUUGUUUGUUUAUGCUCCUUUUUGGGCACACUUACUUG ...(((((((((((.......)))))..((((((.(((...(....)...))).)))).((((.(((......))).)))).........))))))))....... (-18.17 = -18.93 + 0.75)

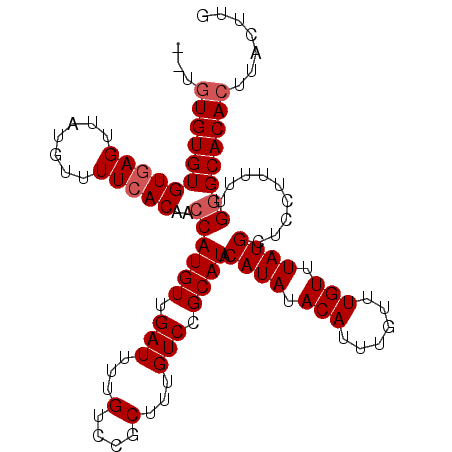
| Location | 6,572,095 – 6,572,198 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -18.95 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6572095 103 - 22407834 CAAGUAAGUGUGCCCAAAAAGGAGCAUAAACAAACAAAUGUAUAUGUAUGCGGACAAAGCGGACAAAAUCAACAUGGUUGUGAAAACAUAACUAACACACAAA-- .......(((((.((........(((((.(((......))).)))))..((.......))))............((((((((....)))))))).)))))...-- ( -22.10) >DroSim_CAF1 46135 103 - 1 CAAGUAAGUGUGCCCAAAAAGGAGCAUAAACAAACAAAUGUAUAUGUAUGCGGACAAAGCGGACAAAAUCAACAUGGUUGUGAAAACAUAACUCACACACACA-- .......(((((.((........(((((.(((......))).)))))..((.......)))).............(((((((....)))))))..)))))...-- ( -20.30) >DroEre_CAF1 45013 105 - 1 CAAGUAAGUGUGCCCAAAAAGGAGCAUAAACAAACAAAUGUAUAUGUAUGCGGACAAAGCGGACAAAAUCAACAUGGUUGUGAAAACAUAACUCACACACACACG ...((..(((((.((........(((((.(((......))).)))))..((.......)))).............(((((((....)))))))..)))))..)). ( -20.50) >DroAna_CAF1 46963 103 - 1 CAAGUAAGUGUGCACAAAGAGAAGCAUAAACAAAUAACUGUAUAUGUAUGCGGACAAAGCGAACAAAAUCAACAUGGUUGUGAAAACAUAACUCACACACACA-- .......(((((........((.(((((.(((......))).))))).(((.......))).......)).....(((((((....)))))))..)))))...-- ( -19.00) >consensus CAAGUAAGUGUGCCCAAAAAGGAGCAUAAACAAACAAAUGUAUAUGUAUGCGGACAAAGCGGACAAAAUCAACAUGGUUGUGAAAACAUAACUCACACACACA__ .......(((((.((........(((((.(((......))).)))))..((.......)))).............(((((((....)))))))..)))))..... (-18.95 = -19.20 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:24 2006