
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
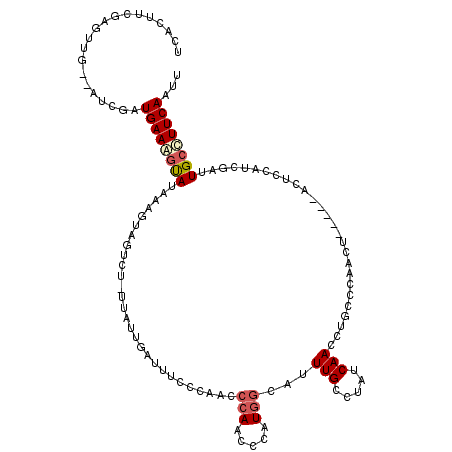
| Location | 6,548,608 – 6,548,722 |
| Length | 114 |
| Max. P | 0.994978 |

| Location | 6,548,608 – 6,548,722 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -4.34 |
| Energy contribution | -4.94 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6548608 114 + 22407834 UCACUUUGAGUUGCAAUCGAUGAAGUUAUAAAGUAGUCUAUUAUUGAUUUUCCAACCCAAUCCAUGGCAUUUGCCUAUCAACCUGCCCAACU-----ACUCCAUCGAUUGCCUUCAAUU .....(((((..((((((((((.(((......((((.......(((......)))..........(((....))).......))))......-----))).)))))))))).))))).. ( -27.10) >DroSec_CAF1 20602 114 + 1 UCACUUCGAGUUGCAAUCGAUGAAAGUAUAAAGUAGUCAGUUAUUGAGUUCCCAACCCAAUCCAUGGCAUUUGCCUAUCAACCUGCCCAACU-----ACUCCAUCGAUUGCUUUCAUUU .......(((..((((((((((..((((....((((......((((.((.....)).))))....(((....))).......)))).....)-----))).)))))))))).))).... ( -26.30) >DroSim_CAF1 22521 114 + 1 UCACUUCGAGUUGAUAUCGAUGAAAGCAUAAAGUAGUCUCUUAUUGAUUUCCCAACCCAACCAAUGAUAUUUGCCUAUCAACCUGCCCAACU-----ACUCCAUCGAUUGCCUUCAAUU ........((((((.(((((((.........((((((......(((......))).........(((((......))))).........)))-----))).))))))).....)))))) ( -15.60) >DroEre_CAF1 21204 96 + 1 GUGCUUCGAC----------UGAAGGUAGAAAGUAGU-------------CCAAGCCCAUCCCAUGGGAUUUGCCUAUCAACCUGUCCAACUCCCGUACUGCGUCGAUUGCCUUCAAUU ..........----------(((((((((...(((((-------------.............((((((.(((..((......))..))).))))))))))).....)))))))))... ( -21.41) >DroYak_CAF1 21659 109 + 1 UUUCUGUGAC----------UGAAAGCAUAAAGUAGUCGUUUAUUGAUUUCCAAGCCCAAAGCAUUGGACUUGUCUAUCAACCUGUCCAAAUCCGGCACUCCAUCGAUUGCCUUCAAUU ....((.(((----------.((..((.....))..)).....(((((...((((.((((....)))).))))...)))))...))))).....((((.((....)).))))....... ( -21.00) >consensus UCACUUCGAGUUG__AUCGAUGAAAGUAUAAAGUAGUCU_UUAUUGAUUUCCCAACCCAACCCAUGGCAUUUGCCUAUCAACCUGCCCAACU_____ACUCCAUCGAUUGCCUUCAAUU ....................((((((((............................(((.....)))...(((.....)))...........................))))))))... ( -4.34 = -4.94 + 0.60)

| Location | 6,548,608 – 6,548,722 |
|---|---|
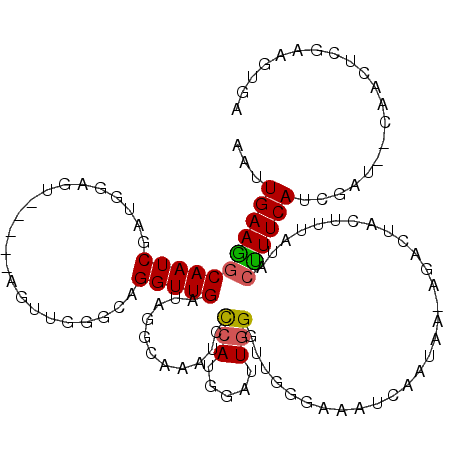
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -7.43 |
| Energy contribution | -7.27 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.23 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6548608 114 - 22407834 AAUUGAAGGCAAUCGAUGGAGU-----AGUUGGGCAGGUUGAUAGGCAAAUGCCAUGGAUUGGGUUGGAAAAUCAAUAAUAGACUACUUUAUAACUUCAUCGAUUGCAACUCAAAGUGA ..((((..((((((((((((((-----.((..((.(((((.((.(((....))))).)))..((((....)))).........)).))..)).))))))))))))))...))))..... ( -36.40) >DroSec_CAF1 20602 114 - 1 AAAUGAAAGCAAUCGAUGGAGU-----AGUUGGGCAGGUUGAUAGGCAAAUGCCAUGGAUUGGGUUGGGAACUCAAUAACUGACUACUUUAUACUUUCAUCGAUUGCAACUCGAAGUGA ........((((((((((((((-----(((..(...........(((....)))....(((((((.....)))))))..)..)))))))........))))))))))............ ( -36.50) >DroSim_CAF1 22521 114 - 1 AAUUGAAGGCAAUCGAUGGAGU-----AGUUGGGCAGGUUGAUAGGCAAAUAUCAUUGGUUGGGUUGGGAAAUCAAUAAGAGACUACUUUAUGCUUUCAUCGAUAUCAACUCGAAGUGA ...((((((((.....((((((-----((((...(..((((((...(((...(((.....))).)))....))))))..).))))))))))))))))))((((.......))))..... ( -27.60) >DroEre_CAF1 21204 96 - 1 AAUUGAAGGCAAUCGACGCAGUACGGGAGUUGGACAGGUUGAUAGGCAAAUCCCAUGGGAUGGGCUUGG-------------ACUACUUUCUACCUUCA----------GUCGAAGCAC ........((..(((((........((((.((((.((((...(((((..((((....))))..))))).-------------...)))))))).)))).----------))))).)).. ( -28.30) >DroYak_CAF1 21659 109 - 1 AAUUGAAGGCAAUCGAUGGAGUGCCGGAUUUGGACAGGUUGAUAGACAAGUCCAAUGCUUUGGGCUUGGAAAUCAAUAAACGACUACUUUAUGCUUUCA----------GUCACAGAAA .((((((((((.....((((((((((....)))....((((((...(((((((((....)))))))))...)))))).......)))))))))))))))----------))........ ( -32.30) >consensus AAUUGAAGGCAAUCGAUGGAGU_____AGUUGGGCAGGUUGAUAGGCAAAUCCCAUGGAUUGGGUUGGGAAAUCAAUAA_AGACUACUUUAUACUUUCAUCGAU__CAACUCGAAGUGA ...(((((((((((......................)))))...........(((.....)))..............................)))))).................... ( -7.43 = -7.27 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:13 2006